Abstract
Human SHPRH gene is located at the 6q24 chromosomal region, and loss of heterozygosity in this region is seen in a wide variety of cancers. SHPRH is a member of the SWI/SNF family of ATPases/helicases, and it possesses a C3HC4 RING motif characteristic of ubiquitin ligase proteins. In both of these features, SHPRH resembles the yeast Rad5 protein, which, together with Mms2–Ubc13, promotes replication through DNA lesions via an error-free postreplicational repair pathway. Genetic evidence in yeast has indicated a role for Rad5 as a ubiquitin ligase in mediating the Mms2–Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Here we show that SHPRH is a functional homolog of Rad5. Similar to Rad5, SHPRH physically interacts with the Rad6–Rad18 and Mms2–Ubc13 complexes, and we show that SHPRH protein is a ubiquitin ligase indispensable for Mms2–Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Based on these observations, we predict a role for SHPRH in promoting error-free replication through DNA lesions. Such a role for SHPRH is consistent with the observation that this gene is mutated in a number of cancer cell lines, including those from melanomas and ovarian cancers, which raises the strong possibility that SHPRH function is an important deterrent to mutagenesis and carcinogenesis in humans.
Keywords: postreplication repair, translesion synthesis, tumor suppressor
Genetic and biochemical studies in the yeast Saccharomyces cerevisiae have been instrumental in yielding an understanding of the pathways and mechanisms that eukaryotic cells employ to rescue the replication fork stalled at DNA lesion sites. Unless rescued in a timely and orderly fashion, the stalling of replication can lead to DNA strand breaks and result in chromosome rearrangements and enhanced rates of carcinogenesis.
The Rad6–Rad18 ubiquitin-conjugating complex of yeast (1, 2) governs at least three alternative pathways that promote replication through DNA lesions: DNA polymerase (Pol) η and Polζ-dependent translesion synthesis (TLS), and an Mms2–Ubc13–Rad5-dependent error-free postreplicational repair (PRR) pathway (3). Polη, for example, promotes efficient and relatively error-free synthesis through UV-induced cyclobutane pyrimidine dimers (4); consequently, inactivation of Polη in both yeast and humans confers enhanced UV mutagenesis (5, 6) and in humans causes the variant form of xeroderma pigmentosum, a cancer-prone syndrome (7, 8). Polζ promotes lesion bypass primarily via its role as an extender, wherein, after the insertion of a nucleotide opposite the DNA lesion by another polymerase, Polζ performs the extension of the nascent primer terminus (3, 9). The Mms2–Ubc13–Rad5-dependent pathway promotes the repair of discontinuities that form in the newly synthesized strand opposite from DNA lesions (10). Although the mechanism by which the Rad5 pathway operates is not known, it likely utilizes a template switching mechanism, wherein the newly synthesized daughter strand of the undamaged complementary sequence is used as the template for bypassing the lesion (10, 11).
Rad5, a member of the SWI/SNF family of ATPases (12), exhibits a DNA-dependent ATPase activity (13), and it also has a C3HC4 RING motif, characteristic of ubiquitin ligases (14, 15). Ubiquitin ligases promote the protein ubiquitylation reaction by binding the cognate ubiquitin-conjugating (UBC) enzyme as well as the protein substrate and by positioning them optimally for efficient ubiquitin conjugation to occur (16). As expected for a ubiquitin ligase, Rad5 physically associates with the Mms2–Ubc13 complex via Ubc13, and it also interacts with the Rad18 subunit of the Rad6–Rad18 complex (17). Both the ATPase and ubiquitin ligase activities of Rad5 are essential for PRR, as the repair of discontinuities formed in the newly synthesized DNA in UV irradiated cells becomes as highly impaired in the rad5 mutants defective in either of these functions as in the rad5Δ mutant (18).
In DNA damaged yeast cells, proliferating cell nuclear antigen (PCNA) becomes monoubiquitylated at lysine-164 by Rad6–Rad18; subsequently, this lysine residue is polyubiquitylated via a lysine-63-linked ubiquitin chain in an Mms2–Ubc13–Rad5-dependent manner (19). Although the biochemical evidence is lacking, genetic evidence in yeast is consistent with a role of Rad5 acting as a ubiquitin ligase in Mms2–Ubc13-dependent polyubiquitylation of PCNA (17, 19). All three Rad6–Rad18-dependent lesion bypass pathways are rendered inactive in the absence of PCNA ubiquitylation (19–21). Monoubiquitylation of PCNA is necessary for TLS by Polη and Poζ, and polyubiquitylated PCNA activates the Rad5-dependent PRR pathway. Because PRR becomes as highly defective in the K164R (pol30–119) mutant of PCNA as in the rad5Δ and mms2Δ mutants (20), PCNA polyubiquitylation is a necessary prerequisite for the activation of this PRR pathway.
The various elements of the Rad6–Rad18 pathway have been conserved in higher eukaryotes, including in humans. Also, similar to that in yeast, PCNA is monoubiquitylated and then polyubiquitylated via lysine-63-linked ubiquitin chains in response to treatment with DNA damaging agents (22, 23). Although for humans the various TLS polymerases that promote lesion bypass in a Rad6–Rad18-dependent manner have been described (3) and the Mms2–Ubc13 complex has been identified (24), the evidence for a Rad5 counterpart has been lacking thus far.
Similar to yeast Rad5, human SHPRH is a member of the SWI/SNF family of ATPases, and as in Rad5, SHPRH harbors a C3HC4-type RING-finger motif which is embedded in between the conserved SWI/SNF helicase domains (25). The human SHPRH gene is located in the chromosomal region 6q24, and loss of heterozygosity of this region has been observed in a number of cancers, including melanoma, ovarian cancer, breast cancer, hepatocellular carcinoma, cervical cancer, pancreatic cancer, and endodermal sinus tumors. Furthermore, sequence analyses of SHPRH from a number of tumor cell lines have revealed a number of mutations in this gene that are not present in normal chromosomes. Thus, a tumor suppressor role for SHPRH is indicated in human cells (25).
Here we examine the possibility that SHPRH is a functional homolog of yeast Rad5. We provide evidence that similar to Rad5, SHPRH physically interacts with the human Rad6–Rad18 and Mms2–Ubc13 protein complexes, and importantly, we show that it exhibits an ubiquitin ligase activity and mediates Mms2–Ubc13-dependent polyubiquitylation of PCNA. Thus, SHPRH is a functional homolog of Rad5.
Results
Homology of Human SHPRH and Yeast Rad5.
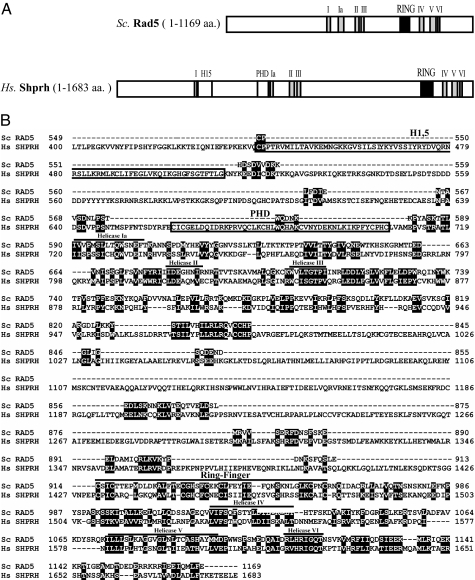
Sequence alignment of human SHPRH and yeast Rad5 proteins reveals a 26% amino acid identity and 37% similarity (Fig. 1). Most of the similarity between yeast Rad5 and human SHPRH is restricted to conserved domains shown schematically in Fig. 1A. Both proteins have a C3HC4-type RING domain and the seven conserved SWI/SNF helicase domains. Importantly, in both proteins the RING domain is embedded uniquely in between the helicase domains (Fig. 1A). In addition to these highly conserved domains, SHPRH has sequence motifs that are not present in Rad5. One such domain is the PHD-finger containing a C4HC3-finger, which is capable of binding zinc ions similar to the RING-finger domain. The other is the linker histone domain that is characteristic of histones of the H1 and H5 families and is termed the H1,5 domain (Fig. 1B).
Fig. 1.
Sequence homology of human SHPRH and yeast Rad5 proteins. (A) Schematic representation of SHPRH and Rad5 proteins. Relative positions of the conserved sequence motifs are represented by boxes. Both SHPRH and Rad5 contain SWI/SNF helicase motifs and a C3HC4 RING-finger domain, which is embedded in between the helicase motifs. The gray boxes numbered from I to VI indicate the conserved helicase motifs, whereas the conserved RING-finger domain is represented by a black box. Additionally, SHPRH also contains an H1,5 domain found in linker histone 1 and histone 5 families and a PHD domain represented by the C4HC3 motif. (B) Alignment of S. cerevisiae Rad5 and human SHPRH proteins. Residues conserved between Rad5 and SHPRH are represented by black boxes. The helicase motifs and the C3HC4 RING finger motif conserved between the two proteins are indicated. The H1,5 histone linker domain and the PHD domain, which are present only in SHPRH, are denoted by boxes. Because of the absence of any interesting features, the N terminus of the two proteins is not shown.
Purification of Human SHPRH, Rad6–Rad18, and Mms2–Ubc13 Proteins.
We wished to determine whether, similar to yeast Rad5, SHPRH functions as a ubiquitin ligase in promoting Mms2–Ubc13-dependent polyubiquitylation of PCNA. Because Rad5 interacts physically with the Mms2–Ubc13 complex via binding to Ubc13, as well as the Rad6–Rad18 complex via binding Rad18, we first determined whether any evidence for similar physical interactions of SHPRH with the Rad6–Rad18 and Mms2–Ubc13 complexes could be observed.
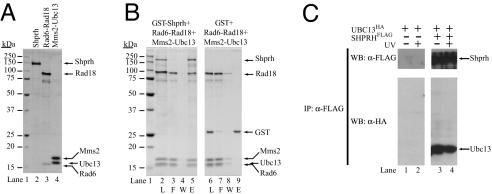
For this purpose, we purified the human SHPRH protein as well as the human Rad6–Rad18 and Mms2–Ubc13 complexes. To facilitate the purification of human SHPRH protein, we fused the SHPRH cDNA downstream of the GST gene and expressed it in a protease-deficient S. cerevisiae strain. The GST–SHPRH protein was purified on glutathione beads, followed by elution of native SHPRH with Pre-Scission protease, which removed the GST tag. The human Rad6–Rad18 complex was purified from yeast cells coexpressing GST–Rad18 together with native Rad6. From glutathione-Sepharose beads, the Rad6–Rad18 complex eluted in a complex. The human Mms2–Ubc13 complex was generated by incubating purified Mms2 together with Ubc13 in a one-to-one molar ratio followed by further purification by gel filtration. As shown in Fig. 2A, the SHPRH protein and the Rad6–Rad18 and Mms2–Ubc13 complexes have been purified to near homogeneity.
Fig. 2.
Physical interaction of SHPRH with Mms2–Ubc13 and Rad6–Rad18. (A) Purity of SHPRH and of Rad6–Rad18 and Mms2–Ubc13 protein complexes. Purified proteins were analyzed on a 12% denaturating polyacrylamide gel and stained with Coomassie blue. Lane 1, molecular mass standards; lane 2, 1 μg of purified SHPRH; lane 3, 1 μg of purified Rad6–Rad18 complex; lane 4, 1 μg of purified Mms2–Ubc13 complex. (B) GST pull-down of purified SHPRH with Rad6–Rad18 and Mms2–Ubc13. GST–SHPRH (3 μg) was mixed with Mms2–Ubc13 (2 μg) and Rad6–Rad18 (6 μg). After incubation, samples were bound to glutathione-Sepharose beads, followed by washings and elution of the bound proteins by glutathione. Aliquots of each sample, taken before addition to the beads (L), from the flow-through fraction (F), from the last washing fraction (W), and from the eluted proteins (E), were analyzed on an SDS/12% polyacrylamide gel stained with Coomassie blue. (C) Immunoprecipitation of human SHPRH in complex with Ubc13 from human cells. HEK293T cells were transiently transfected with plasmids expressing the epitope-tagged HA–Ubc13, alone or together with FLAG-SHPRH proteins as indicated. Thirty-six hours after transfection half of the cells were treated with 42 J/m2 UV, and 5 h later total cellular lysates were prepared. Immunoprecipitations were carried out with anti-FLAG antibodies and subsequently the precipitates were analyzed on Western blots by using anti-FLAG and anti-HA antibodies as indicated.
Physical Interaction of SHPRH with Rad6–Rad18 and Mms2–Ubc13.
To provide evidence for the physical interaction of SHPRH with the Rad6–Rad18 and Mms2–Ubc13 complexes, we added purified Mms2–Ubc13 along with Rad6–Rad18 to the purified GST–SHPRH, and after incubation, we carried out a pull-down assay using glutathione-Sepharose affinity beads. The GST–SHPRH protein was then released from the beads by incubating with glutathione. As shown in Fig. 2B, lane 5, GST–SHPRH eluted together with the Mms2–Ubc13 and Rad6–Rad18 protein complexes, indicating that these proteins formed a multisubunit complex. In control experiments, we verified that GST alone or the glutathione-Sepharose beads did not bind the Mms2–Ubc13 or the Rad6–Rad18 complex (Fig. 2B, lanes 6–9).
We also examined whether SHPRH can form a complex with Mms2–Ubc13 that is stable enough to survive the conditions of gel filtration, a nonequilibrium technique. For this purpose, SHPRH was preincubated with Mms2–Ubc13 followed by gel filtration on a Superdex 200 column. A stable complex between SHPRH and Mms2–Ubc13 was observed, because the elution profile of Mms2–Ubc13 shifted markedly to a higher molecular weight position when it was together with SHPRH compared with the elution position if Mms2–Ubc13 was gel-filtered alone (data not shown).
Next we examined whether SHPRH formed a complex with Ubc13 in vivo and whether UV-irradiation has an effect on this complex formation. HA-UBC13 and FLAG-SHPRH plasmids, or, as a control, HA-UBC13 and empty FLAG vectors, were transfected into HEK293T cells, and this was followed by immunoprecipitation using immobilized anti-FLAG beads. The precipitated proteins were then analyzed for the presence of SHPRH by using anti-FLAG antibody and for the presence of Ubc13 by using anti-HA antibody by Western blot. In parallel experiments, cells were either irradiated or not irradiated with UV-light 5 h before immunoprecipitations. As shown in Fig. 2C, Ubc13 was associated with SHPRH, irrespective of whether the cells were UV irradiated.
Rad6–Rad18 Monoubiquitylates PCNA Encircling DNA.
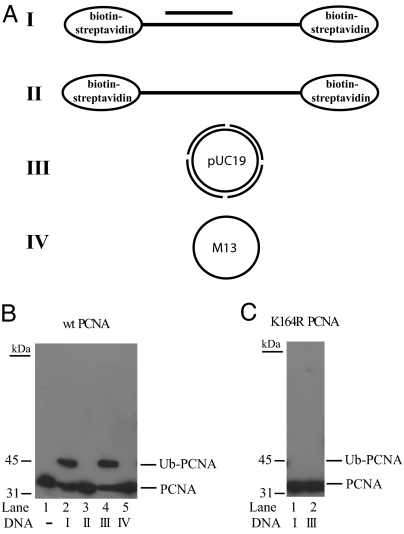
Genetic studies in yeast have indicated that Rad5 is not required for the Rad6–Rad18-dependent monoubiquitylation of PCNA, but the Rad5 function is indispensable for the subsequent polyubiquitylation of monoubiquitylated PCNA (19). Because of the ability of SHPRH to form a physical complex with both the Rad6–Rad18 and Mms2–Ubc13 complexes, we next examined whether similar to Rad5, SHPRH also is not required for Rad6–Rad18-dependent monoubiquitylation of PCNA but is important for its polyubiquitylation. To test for this possibility, we first reconstituted the PCNA monoubiquitylation reaction in vitro using purified Rad6–Rad18. As we and others have shown previously for yeast PCNA (26, 27), incubation of human PCNA together with Rad6–Rad18, ubiquitin, ubiquitin-activating enzyme, and ATP but with no DNA, did not support the ubiquitylation of PCNA (Fig. 3B, lane 1), suggesting that PCNA had to be loaded onto DNA for PCNA ubiquitylation. When PCNA was loaded by RFC onto a 75-nt/31-nt partial heteroduplex DNA, containing biotin-streptavidin complexes at both ends of the 75-nt oligonucleotide to prevent PCNA from sliding off the DNA (Fig. 3A), the Rad6–Rad18 complex efficiently monoubiquitylated PCNA (Fig. 3B, lane 2). Neither single-stranded 75-nt oligonucleotide nor single-stranded M13 DNA (Fig. 3A) was sufficient to promote PCNA monoubiquitylation (Fig. 3B, lanes 3 and 5, respectively), whereas with nicked double-stranded circular plasmid DNA (Fig. 3A), onto which RFC is able to load PCNA (and from which PCNA cannot slide off), we detected efficient PCNA ubiquitylation (Fig. 3B, lane 4). To verify that the conserved lysine-164 residue of PCNA is the ubiquitin acceptor site, we also carried out the ubiquitylation reaction in the presence of a mutant PCNA substrate in which the lysine-164 residue was changed to arginine (Fig. 3C). As expected, this K164R mutant PCNA was not ubiquitylated, thus confirming that the K164 residue is the ubiquitin attachment site in human PCNA. In summary, the human Rad6–Rad18 enzyme complex monoubiquitylates PCNA at its lysine-164 residue, and for this reaction to occur the PCNA must be stably loaded onto the DNA.
Fig. 3.
DNA-dependent monoubiquitylation of PCNA by Rad6–Rad18. (A) Schematic representation of DNAs used in the PCNA ubiquitylation reactions. I, 75/31-nt partial heteroduplex DNA, in which the 75-nt strand contains biotin-streptavidin bound at each end; II, single-stranded 75-nt oligomer containing biotin-streptavidin bound at each end; III, double-stranded pUC19 plasmid DNA nicked enzymatically at four positions; IV, single-stranded M13 DNA. (B) PCNA monoubiquitylation by Rad6–Rad18 in the presence or absence of DNA. The complete reaction mixture contained 50 nM PCNA, 100 nM Rad6–Rad18, 100 nM Uba1, 50 μM ubiquitin, and 10 nM RFC, along with 50 nM oligomeric DNAs (I and II) or 5 nM circular DNA (III and IV). Reactions were carried out in the absence or presence of various DNA samples (I–IV). Monoubiquitylation of PCNA was followed by Western blot using anti-PCNA antibody. (C) K164R mutant PCNA is defective in ubiquitylation. K164R mutant PCNA was subjected to ubiquitylation reaction in the presence of DNA I or III under the conditions described for wild-type PCNA.
SHPRH Is a Ubiquitin Ligase for Mms2–Ubc13-Dependent PCNA Polyubiquitylation.
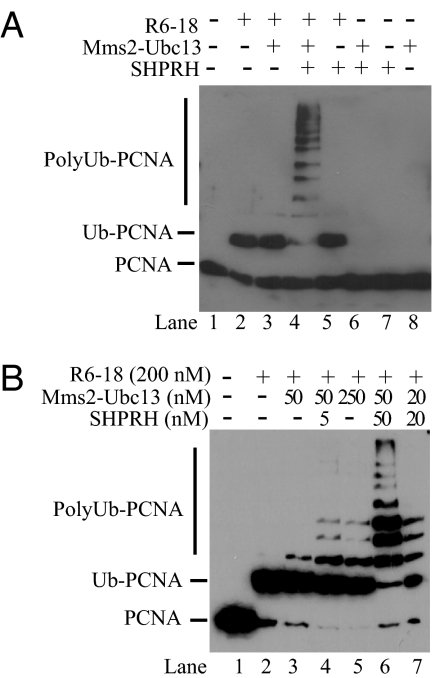
Next, we determined whether PCNA monoubiquitylated at its lysine-164 residue is polyubiquitylated by the Mms2–Ubc13 complex in an SHPRH-dependent manner. For these assays, we first loaded PCNA onto the DNA by RFC, and this was followed by the addition of ubiquitin, Uba1, and various combinations of Rad6–Rad18, Mms2–Ubc13, and SHPRH. Equimolar amounts of Rad6–Rad18, Mms2–Ubc13, and SHPRH were used in these assays. Under the experimental conditions used, the Rad6–Rad18 enzyme complex could monoubiquitylated about fifty percent of PCNA (Fig. 4A, lane 2), and the addition of Mms2–Ubc13 to the reaction did not significantly affect the amount of monoubiquitylated form but resulted in the appearance of a very weak but detectable diubiquitylated form of PCNA (Fig. 4A, lane 3). Strikingly, however, the addition of SHPRH to the Rad6–Rad18 and Mms2–Ubc13-containing reaction resulted in the conversion of almost all of the monoubiquitylated PCNA to polyubiquitylated PCNA species (Fig. 4A, lane 4). In contrast, in the absence of Mms2–Ubc13, SHPRH together with Rad6–Rad18 did not stimulate PCNA polyubiquitylation (Fig. 4A, lane 5). In the absence of Rad6–Rad18, SHPRH together with Mms2–Ubc13 did not support PCNA ubiquitylation (Fig. 4A, lane 6). As expected, neither SHPRH or Mms2–Ubc13 alone show any ubiquitylation activity on PCNA (Fig. 4A, lanes 7 and 8, respectively). Furthermore, the polyubiquitylation of monoubiquitylated PCNA is efficiently catalyzed by Mms2–Ubc13-SHPRH complex in the absence of Rad6–Rad18 (data not presented). Thus, SHPRH is a ubiquitin ligase which acts in concert with the Mms2–Ubc13 and Rad6–Rad18 enzyme complexes in promoting PCNA polyubiquitylation.
Fig. 4.
SHPRH is a ubiquitin ligase that promotes PCNA polyubiquitylation by Mms2–Ubc13. (A) SHPRH is required for PCNA polyubiquitylation. Standard ubiquitylation reaction of PCNA (50 nM) was carried out in the presence or absence of Rad6–Rad18 (100 nM), Mms2–Ubc13 (100 nM), and SHPRH (100 nM). Combinations of these factors were omitted from the reaction as indicated on top. Monoubiquitylation and polyubiquitylation of PCNA were followed by Western blot using anti-PCNA antibody. (B) SHPRH and Mms2–Ubc13 act in a stoichiometric complex. Polyubiquitylation of PCNA (50 nM) was carried out at constant Rad6–Rad18 concentration (200 nM) but at different levels of SHPRH (5–50 nM) and Mms2–Ubc13 (20–250 nM) as indicated.
Next, we determined the optimal stoichiometry of SHPRH and Mms2–Ubc13 for efficient polyubiquitylation of PCNA. The action of SHPRH as a ubiquitin ligase would suggest that it binds to the Mms2–Ubc13 complex in a stoichiometric fashion; and in fact, such a possibility is supported from the interaction studies shown in Fig. 2B, indicating that SHPRH forms a near stoichiometric complex with Mms2–Ubc13. To explore this question further, we carried out PCNA polyubiquitylation assays at different molar ratios of SHPRH and Mms2–Ubc13 (Fig. 4B). For these reactions we added excess Rad6–Rad18 so that almost all of the PCNA was monoubiquitylated (Fig. 4B, lane 2). Importantly, in the absence of SHPRH, Mms2–Ubc13, was unable to carry out efficient polyubiquitylation of PCNA even when it was added at a 5-fold molar excess to PCNA (Fig. 4B, lane 5), whereas addition of even subequimolar amounts of SHPRH enhanced the Mms2–Ubc13-dependent PCNA polyubiquitylation reaction (Fig. 4B, compare lanes 3 and 4) and the stimulation increased proportionally as the concentration of SHPRH was increased (Fig. 4B, compare lanes 4 and 6). Furthermore, the greatest stimulation was observed when SHPRH and Mms2–Ubc13 were present in equimolar amounts (Fig. 4B, lanes 6 and 7). Thus, as for other RING-finger ubiquitin ligases (16), SHPRH affects the activity of Mms2–Ubc13 ubiquitin-conjugating enzyme by forming a stoichiometric complex with it. In summary, our experiments provide strong biochemical evidence that SHPRH is a ubiquitin ligase that promotes PCNA polyubiquitylation in concert with the Mms2–Ubc13 and Rad6–Rad18 enzymes.
Discussion
Here we provide biochemical evidence that human SHPRH is a functional homolog of yeast Rad5 and that it acts as a ubiquitin ligase for promoting Mms2–Ubc13-dependent polyubiquitylation of PCNA. As would be expected for a ubiquitin ligase, SHPRH physically interacts with the Rad6–Rad18 and Mms2–Ubc13 complexes, and a complex containing all these proteins can be purified. In addition, we provide evidence for the association of SHPRH with Ubc13 in human cells. To examine the role of SHPRH in PCNA polyubiquitylation, we reconstituted the Rad6–Rad18-dependent monoubiquitylation of PCNA at its lysine-164 residue and show that Mms2–Ubc13-dependent polyubiquitylation of this PCNA residue requires the SHPRH protein. Furthermore, and as would be expected for a ubiquitin ligase, SHPRH acts in a stoichiometric fashion with Mms2–Ubc13 in supporting the polyubiquitylation reaction.
Genetic studies in yeast have indicated a prominent role for Rad5 and Mms2–Ubc13 in promoting replication through DNA lesions. Although the mechanism by which this pathway operates is not known, it likely uses a copy-choice type of synthesis, in which the information to fill in the gap opposite from the DNA lesion is obtained by copying the newly synthesized strand of the sister duplex. The ubiquitin ligase function of Rad5 performs an indispensable role in PRR by promoting the Mms2–Ubc13-dependent polyubiquitylation of PCNA. Because PRR is equally defective in the rad5Δ, mms2Δ, and the K164R mutant of PCNA, PCNA polyubiquitylation is obligatory for the PRR process to ensue. The SWI/SNF ATPase activity of Rad5 is also critical for PRR, because the inactivation of this function causes the same high degree of PRR defect as that conferred upon inactivating its ubiquitin ligase function.
The Rad5–Mms2–Ubc13-dependent PRR pathway promotes lesion bypass in an error-free manner. For UV lesion, in addition to this pathway, Polη also contributes to error-free bypass via its role in promoting error-free TLS through cyclobutane pyrimidine dimers. Consequently, the frequency of UV induced mutations rises dramatically when both the Rad5-dependent PRR and Polη-dependent TLS pathways are inactivated (28, 29). The inactivation of Polη in humans also leads to an increase in the frequency of UV induced mutations and causes the cancer-prone syndrome, the variant form of xeroderma pigmentosum. Also, there is evidence in human cells that inactivation of Mms2 leads to an enhancement of mutation frequencies (30), and that PCNA polyubiquitylation protects human cells against mutations caused by TLS (22). Overall, the evidence for both yeast and human cells is consistent with a role for the Rad5–Mms2–Ubc13 complex in promoting error-free lesion bypass.
We expect SHPRH to resemble Rad5 in promoting error-free lesion bypass. Such a role predicts that inactivation of SHPRH would confer elevated levels of mutagenesis, resulting from the increased dependence on the TLS polymerases for lesion bypass. In this regard, it is of interest to note that loss of heterozygosity at the human chromosomal band region 6q24-q27, where the SHPRH gene is located, has been observed in multiple human malignancies, particularly in melanomas and breast and ovarian cancers. Moreover, sequence analysis of 44 melanoma and ovarian tumor cell lines has identified four mutations in the SHPRH gene, whereas none of these mutations are seen in normal cell lines; three of these mutations were observed in the hemizygous or homozygous state, indicating a complete lack of functional SHPRH protein in these cell lines (25). These observations, taken together with our evidence that SHPRH is a ubiquitin ligase for mediating Mms2–Ubc13-dependent PCNA polyubiquitylation, and the fact that this pathway promotes error-free replication through the DNA lesions, raise the strong possibility that SHPRH function provides an important deterrent to mutagenesis and carcinogenesis in human cells.
Materials and Methods
Cloning of the cDNAs of Human Genes.
The full-length cDNA of SHPRH was generated from three clones containing different parts of the cDNA, KIAA2023 clone (purchased from Kazusa DNA Research Institute, Chiba, Japan) and DKFZp686L17246 and DKFZp686E02163 clones (purchased from RZPD German Resource Center for Genome Research, Berlin, Germany), and also by RT-PCR of mRNA population of 293T cells. The sequences of all cDNAs were verified by sequencing. The full-length cDNA encoding a 1,683-aa protein was cloned in N-terminal fusion with the glutathione S-transferase gene in the yeast GAL-PGK expression vector pBJ842 (31) resulting in plasmid pIL1467. The cDNAs of human MMS2, UBC13, and PCNA genes were PCR-amplified from a human testis cDNA library (Clontech, Mountain View, CA), and the PCR products were cloned into pBJ842, resulting in plasmids pIL1248, pIL1151, and pIL1238, respectively. For expression in mammalian cells, the PCR-amplified cDNAs of UBC13 and SHPRH were cloned into pRK2H and pRK2F, respectively, resulting in plasmids pIL1501 and pIL1500, respectively. The K164R mutation of human PCNA was generated by PCR-based site-directed mutagenesis (Clontech), and the sequence was verified. The DNA fragment carrying the K164R mutation replaced the wild-type PCNA fragment in plasmid pIL1238, resulting in pIL1254. The cDNA of the hRAD6B gene was generated by PCR and cloned into a yeast ADC1 expression vector, resulting in plasmid pIL182.
The cDNA of human RAD18 was cloned into a modified pBJ842 vector, resulting in plasmid pIL1245, and the sequence of the cDNA was confirmed.
Proteins.
Bovine ubiquitin was purchased from Sigma (St. Louis. MO); ubiquitin-activating enzyme (Uba1) (2) and human RFC (32) were purified as described previously. The human GST–SHPRH, GST–Mms2, GST–Ubc13, and the wild-type and K164R mutant GST–PCNA proteins were expressed individually, whereas GST–Rad18 was coexpressed with native Rad6B in a protease-deficient yeast strain, BJ5464. From total yeast protein extracts obtained from 5 g of yeast cells, GST fusion proteins were bound to 100 μl glutathione-Sepharose 4B column as described previously (31), and after extensive washing the glutathione-Sepharose 4B beads were incubated overnight at 4°C with 5 units of PreScission protease, which cleaves GST fusion proteins 7 aa N-terminal from the first methionine, in buffer A containing 40 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40, and 10% glycerol. Alternatively, GST fusion proteins were eluted from the glutathione-Sepharose 4B beads with 500 μl of buffer A containing 10 mM reduced glutathione followed by extensive dialysis against buffer A. The Mms2–Ubc13 enzyme complex was generated by incubating equal amounts of purified Mms2 and Ubc13 proteins in buffer A overnight at 4°C followed by purification of the complex by Superdex 200 gel filtration column (Amersham) in buffer A. The purified Rad6–Rad18 protein sample was examined by similar gel filtration experiment, and, as expected, it was found to contain only the Rad6–Rad18 complex with a one-to-one stoichiometry of the two proteins and no free Rad6 or Rad18 proteins. Finally, protein samples were concentrated to 1 μg/μl by using a Microcon 30 (Millipore, Billerica, MA), and the purified proteins were aliquoted and frozen at −70°C.
GST Pull-Down Assay.
GST pull-down assays were carried out by using a protocol described previously (33). Briefly, purified GST–SHPRH protein (3 μg) was incubated with Mms2–Ubc13 (2 μg) and Rad6–Rad18 (6 μg) in 40 μl of buffer A but containing 75 mM NaCl at 4°C for 60 min and then for 10 min at 25°C. To such a mixture, 20 μl of glutathione-Sepharose beads were added and further incubated for 1 h with constant rocking at 4°C. The beads were spun down, and the unbound protein was collected. The beads were then thoroughly washed three times with 10 volumes of buffer A. Finally, the bound proteins were eluted with 60 μl of buffer A containing 10 mM reduced glutathione. Various fractions were resolved on a 12% SDS/polyacrylamide gel, followed by Coomassie blue R-250 staining. As a control reaction, the above GST pull-down assay was carried out, but by using GST (1 μg) instead of GST–SHPRH.
Cell Culture and Transfection.
HEK293T cells were cultured in DMEM supplemented with 10% FCS and antibiotics. Cells were transfected with Exgene 500 (Fermentas) according to the manufacturer's instruction.
Western Blot Analyses and Immunoprecipitation.
Cells were lysed in a buffer containing 10 mM Tris·HCl (pH 7.6), 1 mM EDTA, 400 mM NaCl, 10% glycerol, and l mM DTT. The lysis buffer was supplemented with Complete protease inhibitor mix (Roche Molecular Biochemicals) before use. To reduce viscosity, the cell lysates were sonicated briefly on ice. After sonication, the lysates were clarified by centrifugation for 20 min at 4°C at 25,000 × g. Immunoprecipitations were carried out from ≈500 μg of cell extracts. The cell extracts were diluted with an equal volume of dilution buffer (same as the lysis buffer, but it contains no NaCl and the glycerol concentration was increased to 20%). FLAG-tagged proteins were immunoprecipitated with immobilized M2 FLAG antibodies (Sigma). The precipitated proteins and the input lysates were analyzed by Western blot by using the 3F10 HA and the M2 FLAG antibodies (Sigma).
Ubiquitylation Reactions.
A standard in vitro ubiquitylation reaction (10 μl) was carried out in P0 buffer (40 mM Tris·HCl, pH 7.5/8 mM MgCl2/100 μg/ml BSA/10% glycerol/100 μM ATP) in the presence of 100 nM Uba1, 50 μM ubiquitin, 10 nM RFC, and 50 nM 75/31-nt partial heteroduplex DNA containing biotin-streptavidin at each end of the 75-nt oligomer at 30°C for 60 min. As indicated in the figures and legends, ubiquitylation reactions of PCNA (50 nM) were initiated by adding Rad6–Rad18 (100–200 nM), Mms2–Ubc13 (20–250 nM), and SHPRH (5–100 nM) at various concentrations and in combinations indicated. Samples containing unmodified and ubiquitylated PCNA were separated on 12% denaturing polyacrylamide gel and visualized by Western blot by using anti-PCNA antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Typically, in ubiquitylation reactions a 75/31-nt partial heteroduplex linear DNA molecule shown in Fig. 3A was used, which was generated by annealing a 75-nt oligomer, 5′-AGC TAC CAT GCC TGC CTC AAG AAT TCC CAT TAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′, containing one biotin molecule at each end to a 31-nt oligomer primer, 5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG GCA T-3′. Before the reaction we preincubated the above biotin-containing 75/31-nt partial heteroduplex (10 pmol) with streptavidin (20 μg) in 25 μl of reaction buffer. In experiments shown in Fig. 3, the single-stranded 75-nt DNA alone, a single-stranded M13 DNA, and a double-stranded pUC19 plasmid nicked by BstNBI enzyme (New England BioLabs) were also used.
Acknowledgments
We thank Masaru Yamaizumi (Kumamoto University, Kumamoto, Japan) for the human RAD18 gene and Katalin Kovacs for technical assistance. This work was supported by a Wellcome Trust International Senior Research Fellowship, Hungarian Science Foundation Grants OTKA T043354 and OTKA 061921, Marie Curie International Reintegration Grant 022345, Howard Hughes Medical Institute Grant 55005612, Hungarian National Office for Research and Technology Grant KFKT-1-2006-0010, National Institute on Environmental Health Sciences Grant ES012411, and National Institutes of Health Grant GM38559.
Abbreviations
- PCNA
proliferating cell nuclear antigen
- PRR
postreplicational repair
- TLS
translesion synthesis
- Pol
DNA polymerase.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 2.Bailly V, Lauder S, Prakash S, Prakash L. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RE, Prakash S, Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 5.Stary A, Kannouche P, Lehmann AR, Sarasin A. J Biol Chem. 2003;278:18767–18775. doi: 10.1074/jbc.M211838200. [DOI] [PubMed] [Google Scholar]
- 6.Yu S-L, Johnson RE, Prakash S, Prakash L. Mol Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson RE, Kondratick CM, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 8.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 9.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 10.Torres-Ramos C, Prakash S, Prakash L. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Lawrence CW. Proc Natl Acad Sci USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RE, Henderson ST, Petes TD, Prakash S, Bankmann M, Prakash L. Mol Cell Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson RE, Prakash S, Prakash L. J Biol Chem. 1994;269:28259–28262. [PubMed] [Google Scholar]
- 14.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 15.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 17.Ulrich HD, Jentsch S. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangavarapu V, Haracska L, Unk I, Johnson RE, Prakash S, Prakash L. Mol Cell Biol. 2006;26:7783–7790. doi: 10.1128/MCB.01260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 20.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stelter P, Ulrich HD. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 22.Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe K, Tateishi S, Kawasuji M, Tsuirmoto T, Inoue H, Yamaizumi M. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraes TF, Edwards RA, McKenna S, Pastushok L, Xiao W, Glover JNM, Ellison MJ. Nat Struct Biol. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- 25.Sood R, Makalowska I, Galdzicki M, Hu P, Eddings E, Robbins CM, Moses T, Namkoong J, Chen S, Trent JM. Genomics. 2003;82:153–161. doi: 10.1016/s0888-7543(03)00121-6. [DOI] [PubMed] [Google Scholar]
- 26.Garg P, Burgers PM. Proc Natl Acad Sci USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haracska L, Unk I, Prakash L, Prakash S. Proc Natl Acad Sci USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson RE, Prakash S, Prakash L. J Biol Chem. 1999;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 29.McDonald JP, Levine AS, Woodgate R. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Xiao W, McCormick JJ, Maher VM. Proc Natl Acad Sci USA. 2002;99:4459–4464. doi: 10.1073/pnas.062047799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RE, Prakash L, Prakash S. Methods Enzymol. 2006;408:390–407. doi: 10.1016/S0076-6879(06)08024-4. [DOI] [PubMed] [Google Scholar]
- 32.Gibbs E, Kelman Z, Gulbis JM, O'Donnell M, Kuriyan J, Burgers PM, Hurwitz J. J Biol Chem. 1997;272:2373–2381. doi: 10.1074/jbc.272.4.2373. [DOI] [PubMed] [Google Scholar]
- 33.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]