Abstract
Saccharomyces cerevisiae mutants lacking two of the three DNA helicases Sgs1, Srs2, and Rrm3 exhibit slow growth that is suppressed by disrupting homologous recombination. Cells lacking Sgs1 and Rrm3 accumulate gross-chromosomal rearrangements (GCRs) that are suppressed by the DNA damage checkpoint and by homologous recombination-defective mutations. In contrast, rrm3, srs2, and srs2 rrm3 mutants have wild-type GCR rates. GCR types in helicase double mutants include telomere additions, translocations, and broken DNAs healed by a complex process of hairpin-mediated inversion. Spontaneous activation of the Rad53 checkpoint kinase in the rrm3 mutant depends on the Mec3/Rad24 DNA damage sensors and results from activation of the Mec1/Rad9-dependent DNA damage response rather than the Mrc1-dependent replication stress response. Moreover, helicase double mutants accumulate Rad51-dependent Ddc2 foci, indicating the presence of recombination intermediates that are sensed by checkpoints. These findings demonstrate that different nonreplicative helicases function at the interface between replication and repair to maintain genome integrity.
Keywords: checkpoints, genome instability, RRM3, SGS1
In Saccharomyces cerevisiae, the DNA helicases Sgs1, Srs2, and Rrm3 are thought to be important for the maintenance of genome stability. Mutations in any two of these helicase genes cause slow growth that is suppressed by inhibiting homologous recombination (HR). This has been interpreted as suggesting that these helicases constitute a network of partially functionally redundant proteins that act to prevent the deleterious effects of some type of replication error possibly including inherent instability of the replication machinery leading to toxic DNA structures formed by HR proteins (1–5).
Sgs1 is the only RecQ-family, 3′-to-5′ helicase in S. cerevisiae. Sgs1 acts in the coordination between replication and HR, in the suppression of homeologous recombination, in the suppression of crossover products during HR, in S-phase checkpoint activation, and in transcription (6–11). Cells that lack Sgs1 are hyperrecombinogenic and sensitive to DNA-damaging agents such as hydroxyurea and methyl-methanesulfonate, and they accumulate rDNA circles, missegregate chromosomes, and have modestly increased gross-chromosomal rearrangement (GCR) rates (11–15). Mutations in human RecQ-like helicases, such as WRN (16), BLM (17), and RECQ4 (18), cause Werner, Bloom's, and Type-II-Rothmund Thomson syndromes, respectively, characterized by a predisposition to cancer and/or accelerated aging. RecQ-family helicases interact with proteins important for genome stability, such as Rad51 (19) and RPA (20–22), as well as with proteins that function in DNA-damage response pathways (7, 10, 23). Consistent with these findings, sgs1 mutants show incomplete activation of the Rad53 checkpoint kinase in response to DNA-damaging agents (7, 24, 25).
Srs2 is a 3′-to-5′ DNA helicase with similarity to the bacterial Rep and UvrD helicases (26, 27). Srs2 removes Rad51 from single-stranded DNA in vitro, and this is believed to underlie the role of Srs2 as an antirecombinase (28–30). A role for Srs2 in the prevention of aberrant HR is supported by genetic evidence that the hyperrecombination and DNA-damage sensitive phenotypes of srs2 mutants are suppressed by HR defects (31). The physical interaction between Srs2 and Pol32, a structural subunit of DNA polymerase delta, suggests that Srs2 may act during DNA replication (32). Srs2 also is required for proper activation of Rad53 in response to DNA-damaging agents (7, 33), and Srs2 itself is phosphorylated after cells are exposed to methyl-methanesulfonate, hydroxyurea, or UV light; however, the significance of this phosphorylation is unknown (33).
Unlike Sgs1 and Srs2, the Rrm3 helicase has 5′-to-3′ polarity and shares homology throughout its helicase domain with the S. cerevisiae DNA helicase Pif1 (34). Cells lacking Rrm3 exhibit increased recombination at the rDNA locus and at other tandem repeats (35). The physical interaction between Rrm3 and the replication protein proliferating cell nuclear antigen as well as the role of Rrm3 in preventing genome-wide unscheduled replication fork pausing suggest that Rrm3 acts during DNA replication (34, 36, 37). Roles in replication through telomeric and subtelomeric DNA and in suppression of Ty1 transposition also have been assigned to Rrm3 (38, 39). Interestingly, Rad53 is partially activated in rrm3 mutants (37).
sgs1 rrm3, sgs1 srs2, and srs2 rrm3 double mutants exhibit growth defects that are suppressed by mutations in RAD51, RAD55, or RAD57; this has been interpreted to suggest that the helicase double mutants accumulate toxic HR intermediates, such as Holliday junctions. Holliday junctions are typically thought to form during double-strand break repair (40, 41); however, they also are generated during DNA replication in Escherichia coli as a result of fork pausing, establishing a link between replication and recombination (42). Coupling of replication fork pausing to recombination also is supported by the finding in S. cerevisiae that pausing at the replication-fork barrier in rDNA stimulates recombinational events nearby (43). Spikes of X-shaped molecules identified on 2D gels, which have been interpreted as Holliday junctions because their formation depends on the recombination protein Rad52, are stronger in S-phase than in any other phase of the vegetative cell cycle of S. cerevisiae and may result from repair of replication-related DNA lesions by recombination between sister chromatids (44). X-shaped molecules also are observed in rrm3 mutants, suggesting a link between joint molecule formation and replication fork pausing in the absence of the Rrm3 (39). Rad51-dependent X-shaped DNA also accumulates in methyl-methanesulfonate-treated sgs1 mutants, suggesting that Sgs1 facilitates the resolution of these HR intermediates or prevents their formation (25). However, a relationship between replication-dependent Holliday junctions and other branched, aberrant replication-associated DNA structures or broken DNAs has not yet been established. In addition to dissociating Holliday junctions and G4-DNA in vitro (45, 46), BLM, the human RecQ-like helicase most closely related to Sgs1, also has been shown to melt D-loops formed between oligonucleotides in vitro (47).
In the present study, we have analyzed GCRs and checkpoint activation in recombination and/or checkpoint-defective cells lacking one or more helicases. We found that the helicase double mutants accumulate DNA damage foci and that their frequency is decreased in the absence of Rad51. We observed an increased GCR rate in cells lacking Sgs1 and Rrm3 but not in cells lacking Srs2 and Rrm3, underlining fundamental differences between the roles of these helicases in the maintenance of genome stability. Finally, we present evidence that GCRs in sgs1 rrm3 mutants result from HR and are suppressed by the DNA damage checkpoint response.
Results
GCRs in Helicase Mutants Are Promoted by HR and Suppressed by the DNA Damage Checkpoint.
Cells lacking Sgs1 accumulate GCRs at a moderate rate (11). To gain further insight into the cause of the slow-growth phenotype and decreased viability of helicase double mutants, we examined the accumulation of GCRs in rrm3, srs2, and sgs1 single mutants and in strains with multiple helicase defects. In contrast to an sgs1 mutant, rrm3 and srs2 single mutants and an rrm3 srs2 double mutant did not have increased GCR rates compared with wild type (Table 1). However, the rrm3 sgs1 double mutant had an increased GCR rate compared with the sgs1 single mutant (P = 0.0018). Because helicase double mutants with an additional deletion of RAD51 or RAD55 grow normally, we measured the GCR rates of rrm3 sgs1 rad51 and rrm3 sgs1 rad55 mutants and observed a reduction in GCR rates compared with the rrm3 sgs1 double mutant (P < 0.0001; Table 1). In contrast to a RAD51 deletion, deletion of the DNA damage checkpoint genes MEC3 or RAD24 led to increases in the GCR rate of the sgs1 rrm3 mutant (P = 0.0007 and P = 0.0025, respectively). This finding suggests that aberrant HR initiates most of the GCRs in rrm3 sgs1 mutants, which can be prevented if the DNA damage checkpoints are functional.
Table 1.
Effect of helicase, homologous recombination, and checkpoint defects on the rate of accumulation of GCRs
| Relevant genotype | Wild type | rad51Δ | rad55Δ | mec3Δ | rad24Δ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| RDKY strain | Mutation rate | RDKY strain | Mutation rate | RDKY strain | Mutation rate | RDKY strain | Mutation rate | RDKY strain | Mutation rate | |
| Wild type | 3615 | 3.5 (1) | 5559 | <8 (<2.3) | 5565 | 71 (20) | 5569 | 17 (5) | 5573 | 23 (7) |
| rrm3Δ | 5556 | 14 (4) | 5560 | 13 (4) | 5566 | 119 (34) | 5570 | 18 (5) | 5574 | 18 (5) |
| srs2Δ | 5557 | 2.2 (0.6) | 5561 | 17 (5) | ND | 5571 | 23 (7) | ND | ||
| sgs1Δ | 5558 | 77 (22) | 5562 | 35 (10) | 5567 | 289 (82) | 5572 | 339 (97) | 5575 | 136 (39) |
| rrm3Δsrs2Δ | 5576* | 10 (3) | 5563 | 51 (15) | ND | 5578* | 290 (83) | ND | ||
| rrm3Δsgs1Δ | 5577* | 656 (187) | 5564 | 15 (4) | 5568 | 76 (22) | 5579† | 1,942 (555) | 5580 | 1687 (482) |
Mutation rates (Canr 5-FOAr × 10−10) were calculated as described previously (68) by determining the number of cells that are resistant to canavanine (Canr) and 5-fluoroorotic acid (5-FOAr) due to coinactivation of the CAN1 and URA3 genes on chromosome 5. The numbers in parentheses indicate the increase of the GCR rate relative to wild type. ND, not determined.
*Haploid strains containing mutations of interest were freshly obtained by sporulation of RDKY5576, RDKY5577 and RDKY5578.
†Freshly obtained spores had a GCR rate (8 × 10−7) that was slightly higher, although similar to the haploid strain RDKY5579.
Next we sequenced the breakpoints of the GCRs formed in the rrm3 sgs1 and rrm3 sgs1 mec3 mutants (Table 2; see also Table 3, which is published as supporting information on the PNAS web site). Translocations in the rrm3 sgs1 and rrm3 sgs1 mec3 mutants occurred exclusively between regions with microhomologies (Table 2), consistent with a dependency on HR. Deletion of RRM3 in the sgs1 mutant led to an increase in the rate of translocations with microhomologies (P = 0.0001). Deletion of MEC3 in the sgs1 rrm3 mutant increased the translocation rate further by ≈5-fold (P < 0.0001), which indicates that the DNA damage checkpoint acts in suppressing translocations. In contrast, Mec3 was not required for prevention of de novo telomere additions in the sgs1 rrm3 mutant (P = 0.19). Sequencing of six translocation junctions isolated from the rrm3 sgs1 mec3 mutant revealed one translocation between chromosome V (nucleotide 34074) and chromosome XIV (nucleotide 779339), one translocation between chromosome V (nucleotide 32662) and a Y′ element, and four translocations between the CAN1 gene on chromosome V and a CAN1-related gene, either LYP1 or ALP1, on chromosome XIV (Tables 2 and 3). The detailed investigation of homeology-driven translocations between CAN1 and CAN1-related genes in various sgs1 mutants is reported elsewhere (51). In addition to translocations and telomere additions, we identified a complex rearrangement in the rrm3 sgs1 mec3 mutant that consists of an inversion of 22 nt on chromosome V (nucleotides 34290–34312) followed by a de novo telomere addition (Table 3; see also Fig. 5, which is published as supporting information on the PNAS web site).
Table 2.
Rates of GCR types
| Relevant genotype | RDKY strain | GCR rate |
|||
|---|---|---|---|---|---|
| Telomere addition | Translocation |
Other | |||
| NH | MH | ||||
| Wild type* | 3,615 | 2.9 (14/17) | 0.6 (3/17) | <0.2 (0/17) | <0.2 (0/17) |
| mec3† | 5,569 | 14 (8/10) | <1.7 (0/10) | 3.4 (2/10) | <1.7 (0/10) |
| sgs1‡ | 5,558 | 48 (10/16) | 10 (2/16) | 19 (4/16) | <5 (0/16) |
| rrm3 sgs1 | 5,576 | 358 (6/11) | <60 (0/11) | 239 (4/11) | 60 (1/11)§ |
| rrm3 sgs1 mec3 | 5,579 | 242 (1/8) | <242 (0/8) | 1456 (6/8) | 242 (1/8)¶ |
Mutation rates (Canr 5-FOAr × 10−10) were calculated as described for Table 1. Rates of GCR types were determined by multiplying the fractions of the total that a particular rearrangement represents by the GCR rate of the corresponding strain. Significant differences in GCR type rates were seen between the sgs1 and sgs1 rrm3 mutants for de novo telomere additions (P = 0.0008) and translocations with microhomology (P = 0.0001) and between the rrm3 sgs1 and rrm3 sgs1 mec3 mutants for translocations with microhomology (P < 0.0001). In a PCR screen of 41 sgs1 rrm3 mutants, four GCR rearrangements were found in which the URA3 gene was deleted and the CAN1 ORF was present, suggesting independent inactivation of the two genes. Inactivating point mutations in CAN1 could only be identified in three of the four isolates (E76STOP, Q298H, and DEL760G); the chromosome V breakpoints have not yet been mapped. For the rrm3 sgs1 mec3 mutants, one GCR rearrangement was found in which the wild-type ORF of URA3 is present and a I534V mutation was identified in the CAN1 gene. The last 88 bp of the CAN1 ORF could not be amplified, indicative of a partial gene deletion. There is no evidence that I534V inactivates the Can1 protein. The chromosomal breakpoint was not further identified. MH, microhomology; NH, nonhomology.
†See ref. 49.
‡See ref. 11.
§Translocation (NH) with a 1-nt insertion at the breakpoint.
¶Telomere addition on chromosome V after nucleotides 34312–34317 were deleted and nucleotides 34290–34311 were inverted (see text, Table 3, and Fig. 5 for further description).
Normal Growth of HR-Defective sgs1 rrm3 Mutants Depends on the DNA Damage Checkpoint Sensor Mec3.
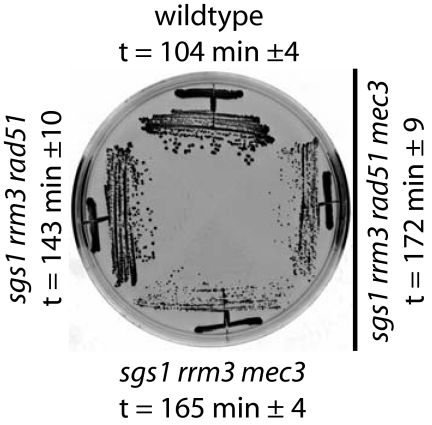
sgs1 rrm3 mutants have a severe growth defect that can be rescued by RAD51 or RAD55 deletions (4, 5, 52). This rescue appears to also depend on the HR protein Rad59 (5), which is thought to promote a Rad52-dependent HR process that requires shorter homologies than Rad51 (53). The rescue of the sgs1 rrm3 slow growth phenotype by a rad51 mutation also depends on the DNA damage checkpoint sensor Mec3 (Fig. 1). sgs1 rrm3 double mutants also have an increased GCR rate that is reduced by deletion of RAD51, and this reduction in the GCR rate was reversed by deletion of MEC3, with the GCR rate of the sgs1 rrm3 rad51 mec3 mutant being equal to the GCR rate of the sgs1 rrm3 mec3 mutant (51), suggesting that normal growth and the low GCR rate of sgs1 rrm3 rad51 mutants require the DNA damage checkpoint.
Fig. 1.
Normal growth of an HR-defective sgs1 rrm3 mutant depends on the DNA-damage checkpoint sensor Mec3. Cells are shown after incubation at 30°C for 2 days on yeast extract/peptone/dextrose media. The doubling time (t) of the sgs1 rrm3 mutant was 267 ± 16 min.
Increased Frequency of Ddc2 Foci in Helicase Mutants Is Rad51-Dependent.
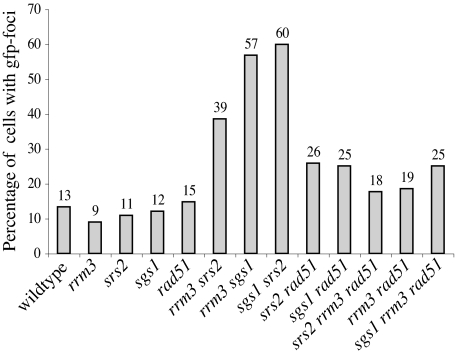
The increased GCR rate seen in sgs1 rrm3 and rrm3 srs2 mutants that lack Rad24 or Mec3 suggests that helicase double mutants suffer spontaneous DNA damage during replication that is sensed by the DNA damage checkpoint, thereby preventing it from engaging in aberrant repair processes. Ddc2, a checkpoint protein that associates with Mec1, relocalizes from a diffuse nuclear distribution into distinct, Mec1-dependent foci after exposure to exogenous DNA-damaging agents or after accumulation of endogenous DNA damage (54–56). To assess checkpoint activation in the sgs1 rrm3 double mutant, we tagged the endogenous copy of DDC2 with GFP and inspected unsynchronized live cells for assembly of Ddc2 foci. Mostly one Ddc2 focus appeared in 9–15% of wild-type cells and helicase or HR single mutants (Fig. 2). One to three Ddc2 foci were found in helicase double mutants, and the percentage of cells with Ddc2 foci increased to 39–60% (Fig. 2). Deletion of RAD51 in these helicase double mutants decreased the number of cells with Ddc2 foci to a level similar to that of HR-deficient helicase single mutants. This finding suggests that HR intermediates that are generated at lesions formed during DNA replication due to lack of Sgs1, Srs2, and Rrm3 are mostly responsible for the increased frequency of Ddc2 foci in helicase double mutants.
Fig. 2.
Ddc2-GFP foci formation at spontaneous DNA damage in helicase double mutants depends on homologous recombination. Quantification of Ddc2-GFP foci in living cells containing helicase mutations and/or a deletion of RAD51. From at least two cell cultures 200–500 cells were analyzed for the presence of foci. Strains were obtained by sporulation of the appropriate diploid strains just before analysis (see Materials and Methods). For images of Ddc2-GFP foci in living cells containing deletions of SGS1, RRM3, SRS2, SGS1, and RRM3; SGS1 and SRS2; or RRM3 and SRS2 (see Fig. 6, which is published as supporting information on the PNAS web site).
Rad53 Phosphorylation in the Absence of Rrm3 Depends on the DNA Damage Checkpoint.
Replication forks are known to pause frequently at specific sites throughout the genome of rrm3 mutants (34, 37, 39), and it is thought that this defect contributes to the slow growth phenotype and increased cell death exhibited by rrm3 mutants that also lack Sgs1 or Srs2. Consistent with this, phosphorylation of Rad53 occurs in rrm3 mutants and persists in rrm3 sgs1 and rrm3 sgs1 rad51 mutants (57). This raises the possibility that replication fork stalling in rrm3 mutants, although insufficient to cause genome instability on its own (Table 1), results in genome instability when cellular processes that combat the deleterious effects of stalled replication forks, such as Sgs1-dependent mechanisms, are defective.
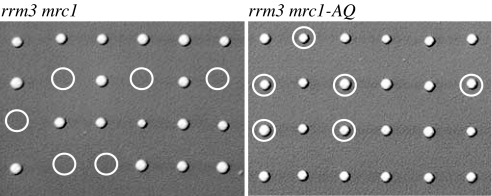
To test the functional significance of Rad53 activation in the absence of Rrm3, we introduced the checkpoint mutations mec3, mec1, rad24, dbp11-1, mrc1, rad9, chk1, dun1, and pds1 into an rrm3 mutant. Only MRC1 was found to be essential for viability of the rrm3 mutant (Fig. 3 and data not shown) and, consistent with a previous report, MEC1 was required for viability at 20°C (37). MRC1 encodes a Claspin-like protein with roles in DNA replication, establishment of sister-chromatid cohesion, and S-phase checkpoint activation (58–61). Mrc1 moves with the replication fork and acts as mediator of replication stress in a Rad9-independent pathway of Rad53 activation and phosphorylation (58–60). Cells lacking Mrc1 are unable to fully activate Rad53 in response to replication stress caused by hydroxyurea treatment, whereas they are fully capable of activating Rad53 in response to DNA damage generated by a cdc13-1 mutation. Because rrm3 mutants suffer from frequent replication fork pausing and exhibit chronic Rad53 phosphorylation, we tested whether the checkpoint function of Mrc1 was required for the survival of rrm3 mutants. An rrm3 mutation was combined with an MRC1 allele (mrc1-AQ) that is proficient for DNA replication but is defective for activation of Rad53 in response to replication stress (59). The rrm3 mrc1-AQ mutant showed normal growth, indicating that the replication function but not the checkpoint function of Mrc1 is essential in rrm3 mutants (Fig. 3).
Fig. 3.
Cells lacking Rrm3 require Mrc1, but not its S-phase checkpoint function, for viability. Tetrads obtained by sporulation of a diploid made by crossing RDKY4714 and RDKY5553 (containing deletions of RRM3 and MRC1, respectively) were dissected and genotyped; spores predicted to contain both rrm3 and mrc1 mutations are absent. Tetrads obtained by sporulation of a diploid made by crossing RDKY4714 and RDKY5554 (containing a deletion of RRM3 and the mrc1-AQ allele, respectively) were dissected and genotyped; rrm3 mrc1-AQ spores are viable. Double mutants are marked by a circle.
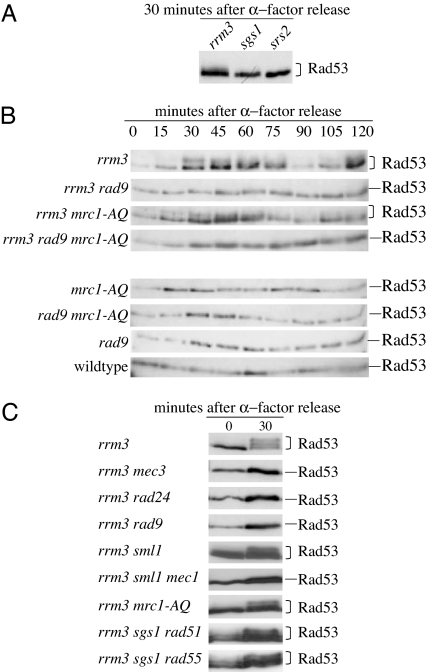
Rad53 is spontaneously phosphorylated in the absence of Rrm3 (37) but not in the absence of Sgs1 or Srs2 (Fig. 4A). Rad53 became phosphorylated during S-phase in an rrm3 mutant, and this phosphorylation was eliminated by the rad9 mutation or by both the rad9 mutation and the mrc1-AQ mutation but not by the mrc1-AQ mutation alone (Fig. 4B). Rad53 phosphorylation also depended on the checkpoint kinase Mec1 and the DNA damage sensors Mec3 and Rad24 (Fig. 4C). These observations suggest that unscheduled fork pausing by itself is not the major source of Rad53 phosphorylation in rrm3 mutants. Instead, these results indicate that Rad53 phosphorylation in rrm3 mutants is mediated through the Mec3/Mec1/Rad9-dependent DNA-damage response pathway and suggests that Rad53 is activated in response to exposure of large single-stranded regions at paused replication forks in rrm3 mutants and/or the conversion of paused replication forks to double-strand breaks. Rad53 was also phosphorylated in rrm3 sgs1 rad51 and rrm3 sgs1 rad55 mutants (Fig. 4C), indicating that recombination intermediates are not the major activator of the DNA damage response in these mutants.
Fig. 4.
Rad53 phosphorylation in rrm3 mutants depends on Mec3, Rad24, Rad9, and Mec1, but not on the S-phase checkpoint function of Mrc1. (A) Rad53 is spontaneously phosphorylated in rrm3 mutants but not in sgs1 or srs2 mutants. Cells were arrested in G1 phase with α-factor released from arrest by transfer into fresh media. Aliquots were harvested just before release from α-factor arrest (time point 0) and after 30 min of continued incubation in fresh media (time point 30). (B) Rad53 phosphorylation in rrm3 mutants is cell-cycle-dependent and Rad9-dependent but does not require the S-phase checkpoint function of Mrc1. Log-phase cultures were released from G1 arrest (time point 0), and aliquots were removed every 15 min for 2 h. Rad53 protein was detected on Western blots by using polyclonal Rad53 antibody (serum JDI47). (C) Rad53 phosphorylation in rrm3 mutants also depends on the checkpoint sensors Rad24 and Mec3 and the checkpoint kinase Mec1. Rad53 phosphorylation in the rrm3 sgs1 mutant is not abolished by a rad51 or rad55 mutation, which rescue the severe, slow-growth phenotype of the rrm3 sgs1 mutant. Cells were treated as in A, and Rad53 protein was detected on Western blots with monoclonal Rad53 antibody (serum EL7).
Similar to the results obtained with mec3 and rad24 mutations (Table 1), the rrm3 rad9 double mutant had essentially the same GCR rate as the rrm3 and rad9 single mutants (3.7 × 10−9, 1.5 × 10−9, and 2 × 10−9, respectively); these results suggest that neither the DNA damage checkpoint nor Rad53 activation accounts for the low GCR rate of rrm3 mutants. Although the amount of DNA damage in rrm3 mutants is sufficiently high to activate Rad53, DNA-damage foci in rrm3 mutants were rare. This may suggest that DNA damage is very efficiently repaired in rrm3 mutants, in part as a result of Rad53-dependent checkpoint activation, so that the threshold of total DNA-damage required for the formation of microscopically detectable Ddc2 foci is rarely reached in cells lacking Rrm3. It should also be noted that the level of Rad53 phosphorylation seen in rrm3 mutants is not as extensive as seen when cells are treated with agents like hydroxyurea or methyl-methanesulfonate that result in full checkpoint activation.
Discussion
In this study, we provide evidence for distinct roles for Sgs1, Srs2, and Rrm3 in the prevention of DNA damage and genome instability. Whereas sgs1 mutants have a moderately increased GCR rate, rrm3 and srs2 mutants have wild-type levels of GCRs. Rrm3 is required in the absence of Sgs1, but not in the absence of Srs2, to prevent GCRs, such as de novo telomere additions and translocations. The increased GCR rate exhibited by rrm3 sgs1 mutants is suppressed by HR-defective mutations but is elevated by mec3 and rad24 mutations. In contrast, HR-defective mutations did not affect the GCR rate of the srs2 rrm3 mutant but deletion of MEC3 led to an increase in the GCR rate, although the resulting GCR rate was much less than that of the mec3 rrm3 sgs1 triple mutant. These differences highlight the distinct roles of Sgs1, Rrm3, and Srs2 in safeguarding genome stability. The different GCR rates may be due to a higher incidence of DNA lesions in the absence of Sgs1 and Rrm3 but not in the absence of Srs2. The lower GCR rate of the rrm3 mutant as compared with the sgs1 mutant, despite the fact that replication forks pause frequently in rrm3 mutants, could be explained by a more random distribution of DNA lesions in the absence of Sgs1, whereas lesions in the rrm3 mutant may be more region-specific (rDNA, subtelomeric DNA, Ty1 elements, and protein-bound sequences) and thus may not occur frequently in the region of chromosome V studied in our GCR assay. Alternatively, it is possible that stalled replication forks per se do not lead to genome instability either because they are not a deleterious form of DNA damage or because cells have potent mechanisms that repair them, including processes that require Sgs1. Additionally, the ability of Sgs1 to prevent illegitimate recombination events may prevent GCRs that might occur in srs2, rrm3, and srs2 rrm3 mutants. Alternatively, loss of Srs2, which has the ability to disrupt Rad51-filamants in vitro (28, 29), may cause significant deregulation of HR in the srs2 and srs2 rrm3 mutants, which may prevent the formation of intermediates that lead to viable GCRs.
The synergistic increase in the GCR rate in sgs1 rrm3 mutants compared with the single mutants provides further evidence that Sgs1 and Rrm3 act in parallel to prevent the formation of HR-dependent DNA lesions during DNA replication that can lead to GCRs. For example, Sgs1 may help replication forks pass through sequences that can adopt unusual DNA structures (46, 62), it may be involved in replication fork stabilization (10) and/or it may act on DNA structures such as Holliday junctions and branched DNA structures that form during replication fork pausing to facilitate fork restart (63, 64). Replication fork pausing at specific sites throughout the S. cerevisiae genome in rrm3 mutants indicates that Rrm3 may help the replication fork bypass proteins that are complexed with DNA or may generally act to prevent aberrant DNA structures that could form when replication forks pause (37, 39, 65); however, either the level of fork pausing in rrm3 mutants or the structure of the stalled forks themselves is not sufficient to result in increased levels of GCRs when Sgs1 is functional. It should be noted that the ability of full-length Rrm3 to act as a helicase and the ability of the Rrm3 helicase domain to unwind DNA substrates other than duplex DNA have not yet been tested (39).
The higher frequency of GCRs in sgs1 mutants as compared with rrm3 and srs2 mutants suggests that Sgs1 has a more significant role in the regulation of DNA replication and/or DNA repair than either Rrm3 or Srs2. For example, although HR is increased significantly in both sgs1 and srs2 mutants, recombination between homeologous sequences increases greatly in sgs1 mutants but shows only a very small increase in srs2 mutants (11, 66). Lack of Sgs1 may allow recombination between similar DNA sequences by allowing pairing between homeologous strands as well as by prolonging the existence of secondary DNA structures, which may normally be unwound by Sgs1. Lack of Rrm3, which prevents unscheduled replication fork pausing, may increase the incidence of DNA lesions that can be processed to GCRs under some conditions. Our observation that the Rad9-dependent DNA damage checkpoint pathway, rather than the Mrc1-dependent replication stress pathway, activates Rad53 in the absence of Rrm3 also supports the notion that DNA breaks or single-stranded gaps accumulate in the absence of Rrm3, possibly because of inappropriate or delayed repair of replication forks that pause or stall frequently in this mutant. The lower GCR rates of sgs1, sgs1 rrm3, and sgs1 rrm3 rad51 mutants compared with the corresponding MEC3-defective mutants indicates that Mec3-dependent checkpoint activation suppresses GCRs. That HR mutations suppress GCRs without eliminating the checkpoint response suggests that the DNA damage that causes Rad53 phosphorylation is not the same DNA damage that induces GCRs. Indeed, Rad53 phosphorylation in rrm3 mutants has been associated with impaired replication of tRNA genes, rDNA, telomeres, centromeres, and mating type loci silencers (34, 37, 39), none of which are present at the chromosome V region analyzed by our GCR assay; persistence of this DNA damage may explain Rad53 phosphorylation in sgs1 rrm3 mutants even when chromosome V-specific GCRs are suppressed by HR mutations. Lack of Mec3 or Rad24 may allow cell cycle progression in the presence of unrepaired DNA lesions or during ongoing lesion repair. The combination of these deficiencies may create a unique environment for the formation of homeology-driven translocation events. Indeed, when we mapped and sequenced GCR breakpoints in the sgs1 rrm3 mec3 mutant we primarily found homeology-mediated translocations (51). In addition, we found evidence of a complex rearrangement process that involves exo- and endonucleolytic DNA cleavage, hairpin formation, DNA synthesis, and de novo telomere addition and results in an inverted sequence (Fig. 5), a type of rearrangement found thus far only in t (11, 22) translocations in Ewing's tumors, which exhibit a high degree of genome-wide instability (67).
Our study supports the view that three nonreplicative helicases, Sgs1, Srs2, and Rrm3, act in different ways to control GCRs during DNA replication: by ensuring replication fork progression (Rrm3 and Sgs1); by preventing secondary structure formation and inter- and intrachromosomal recombination between related sequences (Sgs1); and by regulating the formation of recombinogenic Rad51-filaments (Srs2), emphasizing the importance of both the prevention of illegitimate DNA recombination as well as the limitation of DNA recombination per se for genome stability and normal cell proliferation.
Materials and Methods
Plasmids, Yeast Strains, and GCR Rate Measurement.
S. cerevisiae strains used for the determination of mutation rates are derivatives of S288C. Gene deletions in RDKY3615 (MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8, hxt13::URA3), RDKY5027 (MATα ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8, hxt13::URA3), RDKY2664 (MATα ura3-52 trp1Δ63 his3Δ200), RDKY2666 (MATa ura3-52 trp1Δ63 his3Δ200), and their diploid derivatives were generated by HR-mediated integration of PCR fragments according to standard methods. All haploid strains used for the determination of mutation rates were obtained by sporulation of diploids. To minimize the emergence of suppressors, slow-growing haploid strains were freshly obtained by sporulation of the appropriate diploid strain for every experiment. Haploid strains for the analysis of cell-cycle-dependent Rad53 phosphorylation (Fig. 4B) were obtained by sporulation of the diploid strain YPH501 (MATa/α, ura3-52/ura3-52, lys2-801 amber/lys2-801 amber, ade2-101 ochre/ade2-101 ochre, trp1-Δ63/trp1-Δ63, his3-Δ200/his3-Δ200, leu2-Δ1/leu2-Δ1; gift from V. Zakian, Princeton University) modified to contain heterozygous mutations of interest. To construct RDKY5554, a TRP1 cassette was inserted into the PacI site 250 bp downstream of the mrc1-AQ allele in plasmid pAO138 (gift from S. Elledge, Harvard Medical School, Boston, MA) and this mrc1-AQ.TRP1 fragment replaced a URA3 cassette inserted at the MRC1 locus. Successful integration of the mrc1-AQ allele was confirmed by PCR and sequencing of the entire mrc1-AQ allele. Strains expressing GFP-tagged Ddc2 were obtained by sporulation of RDKY5581, RDKY5582, and RDKY5582. Strains containing pds1 or dpb11-1 mutations were grown at room temperature; all other strains were grown at 30°C. Strains and their complete genotypes are listed in Table 4, which is published as supporting information on the PNAS web site. Media for propagating strains have been described previously (48). GCR rate measurements and breakpoint analyses were carried out as described previously (68); statistical significance of differences in GCR rates was evaluated by using the Mann–Whitney test and programs from R. Lowry at Vasser College (http://faculty.vassar.edu/lowry/vsord.html).
Microscopy.
Live cells were imaged by using a spinning disk confocal scanhead (McBain Instruments, Chatsworth, CA) on a Nikon (Tokyo, Japan) TE2000e inverted microscope. Images were acquired using a ×100, 1.4 N.A. Plan Apo objective lens and an Orca ER CCD camera (Hamamatsu Photonics, Hamamatsu, Japan) with 2 × 2 binning. Acquisition parameters, shutters, and focus were controlled by MetaMorph software (Universal Imaging). Five z sections were acquired at 1-μm steps. Figures were prepared in PhotoShop (Adobe Systems, San Jose, CA), and processing parameters were kept constant. The number of cells with GFP foci was determined by analyzing 200–500 cells.
Cell Cycle Arrest and Western Blot Analysis of Rad53 Phosphorylation.
Cells were grown at 30°C in yeast extract/peptone/dextrose media until they reached OD600 ≈ 0.5, synchronized in G1 with α-factor (15 μg/ml) and released from arrest by replacing the media with fresh, prewarmed yeast extract/peptone/dextrose. Incubation was continued at 30°C for 2 h, and aliquots were removed every 15 min (or aliquots were removed at the beginning and end of a 30-min incubation) (Fig. 4C) and then put on ice and adjusted for cell number. Denatured crude extracts were prepared by trichloroacetic acid extraction (69) and separated on 10% polyacrylamide gels for Western blot analysis. Polyclonal and monoclonal Rad53 antibodies were gifts from J. Diffley (Cancer Research UK, London, United Kingdom) and A. Pellicioli (FIRC Institute of Molecular Oncology Foundation, Milan, Italy), respectively.
Supplementary Material
Acknowledgments
We thank Paul Maddox (Ludwig Institute for Cancer Research) for help with confocal microscopy and image analysis; Vincent Pennaneach, Christopher Putnam, Jorrit Enserinck, and Meng-Er Huang for helpful comments on the manuscript; Virginia Zakian (Princeton University, Princeton, NJ) for strain YPH501; Stephen Elledge (Harvard Medical School, Boston, MA) for plasmid pAO138; J. Diffley (Cancer Research UK) for the polyclonal Rad53 antibody JDI47; and A. Pellicioli (FIRC Institute of Molecular Oncology Foundation) for monoclonal Rad53 antibody EL7. This work was supported by National Institutes of Health Grant GM26017 (to R.D.K.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Gangloff S, Soustelle C, Fabre F. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 2.Klein HL. Genetics. 2001;157:557–565. doi: 10.1093/genetics/157.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabre F, Chan A, Heyer WD, Gangloff S. Proc Natl Acad Sci USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt KH, Kolodner RD. Mol Cell Biol. 2004;24:3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres JZ, Schnakenberg SL, Zakian VA. Mol Cell Biol. 2004;24:3198–3212. doi: 10.1128/MCB.24.8.3198-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 7.Frei C, Gasser SM. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 8.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myung K, Datta A, Chen C, Kolodner RD. Nat Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 12.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinclair DA, Guarente L. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 14.Watt PM, Louis EJ, Borts RH, Hickson ID. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 15.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 17.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 18.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Davies SL, Levitt NC, Hickson ID. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 20.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 21.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cobb JA, Bjergbaek L, Gasser SM. FEBS Lett. 2002;529:43–48. doi: 10.1016/s0014-5793(02)03269-6. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 24.Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong L, Klein HL. J Biol Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- 28.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 29.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 30.Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. J Biol Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- 31.Chanet R, Heude M, Adjiri A, Maloisel L, Fabre F. Mol Cell Biol. 1996;16:4782–4789. doi: 10.1128/mcb.16.9.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang ME, de Calignon A, Nicolas A, Galibert F. Curr Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 33.Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivessa AS, Zhou JQ, Zakian VA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 35.Keil RL, McWilliams AD. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt KH, Derry KL, Kolodner RD. J Biol Chem. 2002;277:45331–45337. doi: 10.1074/jbc.M207263200. [DOI] [PubMed] [Google Scholar]
- 37.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 38.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 41.West SC. Philos Trans R Soc London B. 1995;347:21–25. doi: 10.1098/rstb.1995.0004. [DOI] [PubMed] [Google Scholar]
- 42.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou H, Rothstein R. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 45.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. Proc Natl Acad Sci USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun H, Bennett RJ, Maizels N. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 48.Chen C, Umezu K, Kolodner RD. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 49.Myung K, Chen C, Kolodner RD. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 50.Pennaneach V, Kolodner RD. Nat Genet. 2004;36:612–617. doi: 10.1038/ng1359. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt KH, Wu J, Kolodner RD. Mol Cell Biol. 2006;26:5406–5420. doi: 10.1128/MCB.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ooi SL, Shoemaker DD, Boeke JD. Nat Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 53.Ira G, Haber JE. Mol Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melo JA, Cohen J, Toczyski DP. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tercero JA, Longhese MP, Diffley JF. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 56.Lisby M, Barlow JH, Burgess RC, Rothstein R. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 57.Bessler JB, Zakian VA. Genetics. 2004;168:1205–1218. doi: 10.1534/genetics.104.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- 59.Osborn AJ, Elledge SJ. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 61.Xu H, Boone C, Klein HL. Mol Cell Biol. 2004;24:7082–7090. doi: 10.1128/MCB.24.16.7082-7090.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huber MD, Lee DC, Maizels N. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bennett RJ, Keck JL, Wang JC. J Mol Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- 64.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torres JZ, Bessler JB, Zakian VA. Genes Dev. 2004;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spell RM, Jinks-Robertson S. Genetics. 2004;168:1855–1865. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohali A, Avigad S, Cohen IJ, Meller I, Kollender Y, Issakov J, Goshen Y, Yaniv I, Zaizov R. Cancer Genet Cytogenet. 2004;150:50–56. doi: 10.1016/j.cancergencyto.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 68.Schmidt KH, Pennaneach V, Putnam CD, Kolodner RD. Methods Enzymol. 2006;409:462–476. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- 69.Foiani M, Liberi G, Piatti S, Plevani P. In: Eukaryotic DNA Replication: A Practical Approach. Cotterill S, editor. Oxford, UK: Oxford Univ Press; 1999. pp. 185–200. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.