Abstract
Src-like adaptor protein (SLAP) and c-Cbl recently have been shown to cooperate in regulating T cell receptor (TCR) levels in developing T cells. SLAP also is expressed in developing B cells, and its deficiency leads to alterations in B cell receptor (BCR) levels and B cell development. Hence, we hypothesized that SLAP and c-Cbl may cooperate during B cell development to regulate BCR levels. In mice deficient in both SLAP and c-Cbl, we found that B cell development is altered, suggesting that they function through intersecting pathways. To study the mechanism by which SLAP and c-Cbl alter BCR levels, we coexpressed them in a mature mouse B cell line (Bal-17). First we determined that SLAP associates with proximal components of the BCR complex after stimulation and internalization. Coexpression of SLAP and c-Cbl in Bal-17 led to decreased surface and total BCR levels. This decrease in BCR levels depended on intact Src homology 2 (SH2) and C-terminal domains of SLAP. In addition, a mutation in the SH2 domain of SLAP blocked its colocalization with c-Cbl and the BCR complex, whereas deletion of the C terminus did not affect its localization. Last, coexpression of SLAP and c-Cbl altered BCR complex recycling. This alteration in BCR complex recycling depended on enzymatically active c-Cbl and Src family kinases, as well as the intact SH2 and C-terminal domains of SLAP. These data suggest that SLAP has a conserved function in B and T cells by adapting c-Cbl to the antigen-receptor complex and targeting it for degradation.
Keywords: B cell antigen receptor, B cell development, receptor internalization, ubiquitination
The balance of the immune system can be disrupted by intrinsic defects in lymphocyte signaling pathways (1, 2). Understanding the factors that regulate these signaling pathways could lead to greater insight into pathogenesis and/or treatment of diseases such as immunodeficiency, malignancy, and autoimmunity.
Src-like adaptor protein (SLAP) is a protein that regulates lymphocyte signaling pathways (3, 4). The N terminus of SLAP has sequence and domain similarity to Src family kinases, containing Src homology 3 (SH3) and SH2 domains that are able to bind to activated components of the T cell receptor (TCR) complex (5). Instead of the C terminus containing a kinase domain, it has a unique peptide sequence with a poorly defined function.
Expression of SLAP in cell lines identified it as a negative regulator of cellular signaling (5–7). In vivo studies have shown that SLAP deficiency leads to increased TCR expression selectively on double-positive (DP) thymocytes, where SLAP is most highly expressed (8). Previous studies have shown that the increased expression of the TCR complex on DP thymocytes is attributable to decreased intracellular retention of TCRζ and increased recycling (9). Increased recycling of the TCR to the cell surface leads to increased TCR levels and altered thymocyte development (8, 9). These studies support the notion that SLAP regulates TCR levels during a specific stage of T cell development where selection events occur, and its absence leads to increased signaling through the TCR complex, thereby altering T cell development and maturation.
In addition to SLAP being highly expressed in DP thymocytes, SLAP expression was also detected in mouse B cells (10–12). Recent studies on SLAP function in B cells have demonstrated that expression of SLAP is required to optimally regulate B cell receptor (BCR) levels, signaling through the BCR complex, and B cell development (11). SLAP-deficient B cells expressing a transgenic BCR with single antigen specificity had increased BCR levels on immature (IgM+, IgD−) bone marrow B cells and immature (IgM+, CD21+, IgD−) splenic B cells but decreased BCR levels on mature B cells. In addition, the immature splenic B cells had increased tonic signaling, as evidenced by increased intracellular p-ERK levels. In contrast, mature splenic B cells had a reduced calcium flux in response to signaling through the BCR complex as well as decreased expression of activation-induced surface markers (11). These data suggested that SLAP deficiency in immature B cells resulted in increased BCR levels with increased BCR mediated signaling, which subsequently leads to the development of a hyporesponsive pool of mature B cells expressing lower BCR levels.
Insights into how SLAP regulates TCR levels were obtained from studies where SLAP was shown to associate with c-Cbl in vitro (3, 13). Interestingly, mice deficient in c-Cbl, SLAP, or both have similar thymic phenotypes, suggesting that SLAP and c-Cbl may function in the same biochemical pathway (14–16). Mice deficient in c-Cbl have alterations in B cell development with increased numbers of B1 B cells in the peritoneal cavity and increased immature B cell numbers in the spleen (17). In addition, splenic B cells from c-Cbl-deficient mice have slower kinetics of Lyn activation but sustained activation upon BCR stimulation (17). Furthermore, c-Cbl has been shown to have E3 ubiquitin ligase activity for activated receptor protein tyrosine kinases (PTKs), including those coupled to the TCR complex (18–20). c-Cbl is rapidly phosphorylated after TCR activation and interacts with components of the proximal TCR signaling machinery, including the ZAP-70 tyrosine kinase (21, 22).
Recent work in our laboratory has shown that SLAP is required for c-Cbl-dependent down-modulation of the TCR complex in DP thymocytes (16). As a complement to these studies, we wanted to determine whether SLAP could cooperate with c-Cbl to down-modulate the BCR complex in B cells. Here we demonstrate that B cell development is altered in SLAP and c-Cbl doubly deficient mice. In addition, we determined that in a mouse B cell line, SLAP is required for c-Cbl-dependent down-modulation of the BCR complex.
Results
B cell Development Is Altered in SLAP and c-Cbl Doubly Deficient Mice.
To determine the effects of SLAP and c-Cbl deficiency on B cell development, we performed multiparameter flow cytometric analysis on single-cell suspensions from spleen, bone marrow, and peritoneal cavity from SLAP-deficient, c-Cbl-deficient, SLAP and c-Cbl doubly deficient, and WT C57BL/6 (B6) control mice (Fig. 1). Analysis of bone marrow lymphocytes revealed that the SLAP and c-Cbl doubly deficient mice had decreased recirculating B cells (Fr.F) and increased pro-B cells (Fr.E) compared with the SLAP-deficient, c-Cbl-deficient, and B6 controls (Fig. 1A). These findings suggest that SLAP and c-Cbl function through intersecting pathways leading to alterations in B cell development.
Fig. 1.
SLAP and c-Cbl deficiency lead to alterations in B cell development. (A) Analysis of cells from bone marrow from C57BL/6 (B6), c-Cbl-deficient (c-Cbl−/−), SLAP-deficient (SLAP−/−), or SLAP and c-Cbl doubly deficient mice (DKO). (B) Analysis of splenic B cells (B220+) from B6, c-Cbl−/−, SLAP−/−, or DKO mice. (C) Analysis of peritoneal B cells (CD19+) from B6, c-Cbl−/−, SLAP−/−, or DKO mice. (A–C) The 2D contour plots are representative of results obtained from six mice per genotype. (D) Average percentage of B1 B cells in the peritoneal lavage fluid from B6 (light gray bar), SLAP−/− (black bar), c-Cbl−/− (white bar), or DKO (dark gray bar) mice. (E) Average percentage of B2 B cells in the peritoneal lavage fluid from B6 (light gray bar), SLAP−/− (black bar), c-Cbl−/− (white bar), or DKO (dark gray bar) mice. These data are the average percentage of cells determined from six mice per genotype. The asterisks denote statistically significant differences as compared with the B6 controls.
Analysis of splenic B cells revealed an increased percentage of newly formed (NF) immature B cells in the c-Cbl-deficient and SLAP and c-Cbl doubly deficient mice (Fig. 1B). In addition, the transitional B cells (T1) and marginal zone B cells (Mz), as defined by CD21+ and CD23− surface phenotype, are increased in both the c-Cbl-deficient and the SLAP and c-Cbl doubly deficient mice (Fig. 1B). Further analysis of the T1/MZ subset with anti-CD1d antibodies to define MZ B cells revealed that the increase in the T1/Mz subpopulation was attributable to an increase in the percentage of Mz B cells (data not shown).
Peritoneal lavage from c-Cbl and SLAP and c-Cbl doubly deficient mice had increased percentage of CD5+ B1 B cells with a decreased percentage of CD5− B2 cells (Fig. 1C). These alterations in peritoneal B cell composition are most pronounced in the SLAP and c-Cbl doubly deficient mice and are statistically significant when compared with the B6 controls (Fig. 1 D and E). Together, these data suggest that SLAP and c-Cbl deficiency both alter B cell development and that they function in intersecting pathways leading to greater alterations in B cell development than is seen with either deficiency alone, particularly with regard to the steady-state levels of the Fr.F, NF, and B2 B cell subsets.
SLAP Associates with Components of the BCR Antigen Complex.
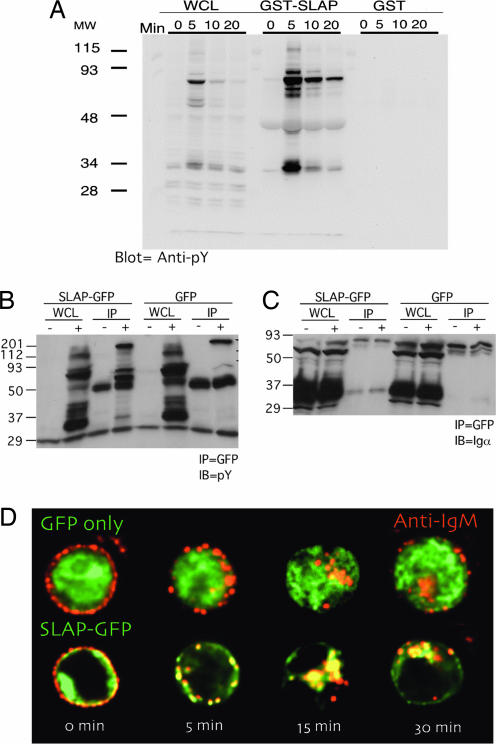
Because strength of signal through the BCR complex influences B cell development and c-Cbl and SLAP both alter BCR levels in vivo, we first wanted to determine whether SLAP associates with components of the BCR complex. In this regard, a fusion of SLAP with GST (GST SLAP) was generated, and binding to phosphoproteins from stimulated BAL-17, a mature B cell line, was determined. GST SLAP specifically associates with proteins that were tyrosine-phosphorylated after BCR stimulation (Fig. 2A). We then determined whether SLAP could specifically associate with proteins that were tyrosine-phosphorylated after BCR stimulation in Bal-17 cells stably expressing a green fluorescent protein (GFP) fusion of SLAP (SLAP-GFP) compared with GFP only as a control. Specifically, SLAP-GFP but not GFP alone could associate with a set of tyrosine-phosphorylated proteins after BCR stimulation (Fig. 2B). Furthermore, SLAP-GFP can constitutively associate with Ig-α, one of signaling chains of the BCR complex (Fig. 2C). In addition, SLAP-GFP in BAL-17 colocalizes with the BCR at the plasma membrane in the basal state. GFP-SLAP also colocalized with the internalized surface BCR after BCR stimulation by anti-IgM F(ab′)2 (Fig. 2D).
Fig. 2.
SLAP associates with the BCR upon stimulation and internalization. (A) Immunoblot of whole-cell lysate (WCL) from BAL-17 cells 0, 5, 10, and 20 min after BCR stimulation. GST pull-downs were performed by using either GST SLAP or GST as a control. Proteins retained during the GST pull-down were detected with an antibody specific for phosphotyrosine. Immunoblots are representative of at least three independent experiments with similar results. (B and C) Immunoblot of WCL from BAL-17 cells stably expressing either GFP or SLAP-GFP 0 and 5 min after BCR stimulation. Coimmunoprecipitations were performed on WCL lysates by using an anti-GFP antibody. Proteins retained during coimmunoprecipitation were detected with an antibody specific for phosphotyrosine (B) or Igα (C). Immunoblots are representative of at least three independent experiments with similar results. (D) Deconvolution microscopy was performed on BAL-17 cells stably expressing either GFP or SLAP-GFP (green). The cells were then fixed 0, 5, 15, and 30 min after BCR stimulation, and IgM (red) and GFP/SLAP-GFP (green) were visualized. Images are representative of at least five independent transfections and at least 20 independent cells captured per transfection.
SLAP and c-Cbl in Bal-17 Cooperatively Down-Regulate Surface and Total BCR Levels, and an Intact SH2 Domain and C Terminus of SLAP Are Required for c-Cbl-Dependent BCR Complex Down-Modulation.
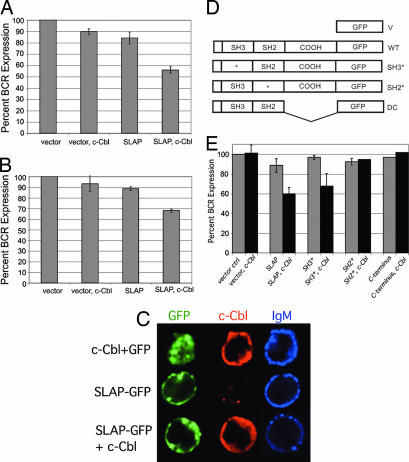
To determine whether SLAP and c-Cbl can modulate BCR levels, constructs expressing both genes were transiently transfected into BAL-17, and surface and total BCR expression was determined by fluorescent-activated cell sorting analysis (FACS). Expression of SLAP-GFP or c-Cbl alone had small effects on BCR surface levels. However, coexpression of SLAP-GFP with c-Cbl led to a 40–50% down-modulation of both surface and total BCR levels (Fig. 3A and B). In addition, we visualized the localization of GFP-SLAP and c-Cbl by using deconvolution microscopy. SLAP-GFP and c-Cbl were present in both the cytoplasm and within vesicles (Fig. 3C). These data demonstrate that SLAP, in conjunction with c-Cbl, regulates BCR levels in BAL-17. The decreased total BCR levels in the transfected cells suggest that SLAP and c-Cbl target the BCR complex for degradation.
Fig. 3.
Expression of SLAP and c-Cbl down-regulates surface and total BCR levels in BAL-17 cells, and both the SH2 and C-terminal domains of SLAP are required for BCR down-modulation. (A) Percentage of surface IgM expression (±SEM) on GFP+ BAL-17 cells transiently transfected with GFP alone, c-Cbl plus GFP, GFP-SLAP, or c-Cbl plus GFP-SLAP. IgM levels were compared with that expressed on GFP-only-transfected cells. The data are the average of at least three independent experiments. (B) Percentage of total IgM expression (±SEM) on GFP+ BAL-17 cells transiently transfected with GFP alone, c-Cbl plus GFP, SLAP-GFP, or c-Cbl plus SLAP-GFP. Total IgM levels were compared with levels expressed on GFP-only-transfected cells. The data are the average of at least three independent experiments. (C) Expression and localization of GFP fluorescence (green), c-Cbl (red), and IgM (blue) in Bal-17 cells that were transiently transfected with GFP or SLAP-GFP with or without c-Cbl and analyzed by deconvolution microscopy. For each experiment >10 cells per transfection condition were analyzed from three independent transfections. (D) SLAP-GFP constructs: GFP only (V), SLAP-GFP (WT), SH3 point mutant (SH3*), SH2 point mutant (SH2*), and deletion of C terminus (DC). (E) Percentage of surface IgM expression (±SEM) on GFP+ BAL-17 cells transiently transfected with GFP-SLAP either alone or with c-Cbl expression constructs. IgM expression was compared with IgM expressed on GFP-only-transfected cells. The data are the average of at least three independent experiments.
We next wanted to establish the domains of SLAP required for BCR down-modulation; a panel of SLAP mutant constructs (Fig. 3D) were cotransfected with c-Cbl, and BCR expression was analyzed by FACS. BCR down-modulation by SLAP and c-Cbl depended on a functional SH2 domain and the presence of the SLAP C terminus (Fig. 3E). In contract, a mutation of the SH3 domain of SLAP had minimal effect on BCR down-modulation (Fig. 3E). These data suggest that BCR down-regulation by SLAP requires an intact SH2 domain as well as the unique C terminus of SLAP.
SLAP with a Mutated SH2 Domain Fails to Localize with the BCR Complex.
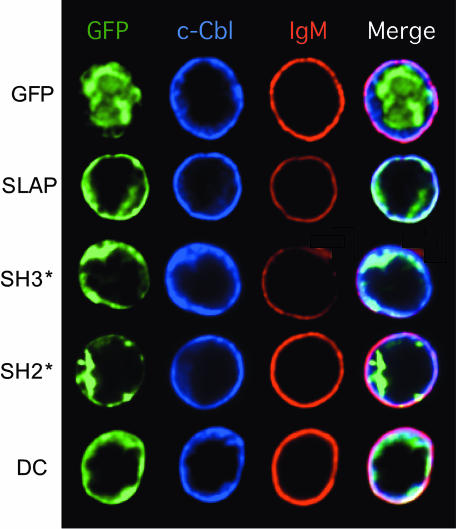
The need for an intact SH2 domain of SLAP for BCR down-modulation and the ability of SLAP to associate with phosphoproteins after BCR stimulation suggest that the SH2 domain of SLAP may be important in targeting it to the BCR complex. To determine whether the SH2 domain of SLAP is required for colocalization with the BCR, we expressed GFP alone (as a control), SLAP-GFP, SH2*-GFP, or SH3*-GFP with c-Cbl in BAL-17 and monitored their localization by using deconvolution microscopy (Fig. 4). GFP alone did not substantially colocalize with either c-Cbl or the BCR. However, c-Cbl was colocalized with the BCR even in the absence of SLAP. Similar to SLAP-GFP, SH3*-GFP colocalized with both c-Cbl and the BCR, supporting the notion that the SH3 domain of SLAP is not required for BCR down-modulation. In contrast, SH2*-GFP had little colocalization with either c-Cbl or the BCR. Lastly, a mutant of SLAP with a truncated C terminus (DC-GFP) also colocalized with c-Cbl and the BCR and localized similarly to SLAP-GFP. However, no BCR down-regulation was noted with the DC-GFP constructs. These data suggest that the SH2 domain of SLAP is important for its association with phosphorylated components of the BCR complex, whereas the C-terminal region is important for BCR down-modulation.
Fig. 4.
The SH2 domain of SLAP is required for its colocalization with IgM in BAL-17. Expression and localization of GFP fluorescence (green), c-Cbl (blue), and IgM (red) in BAL-17 cells transiently transfected with c-Cbl and GFP-SLAP expression constructs and analyzed by deconvolution microscopy. For each experiment >10 cells per transfection condition were analyzed from three independent transfections.
Expression of SLAP and c-Cbl in Bal-17 Cells Leads to Alterations in BCR Recycling.
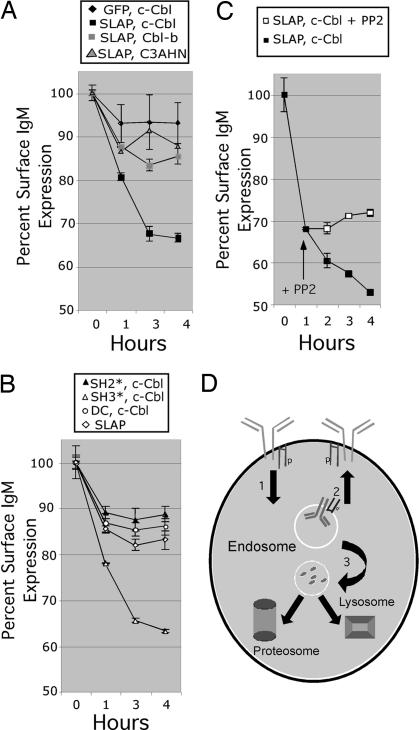
The decreased BCR levels observed upon SLAP and c-Cbl expression could be attributable to alterations in the internalization and/or recycling of the BCR. To determine whether constitutive BCR recycling is altered after SLAP and c-Cbl expression, we developed an assay to monitor the dynamics of BCR recycling in BAL-17 (Fig. 5). Simply, so as not to cross-link the BCR and induce BCR internalization, we used a Fab specific for mouse IgM for these studies. BAL-17 was transiently transfected with the indicated expression constructs for 4 h, labeled on ice with the biotinylated Fab anti-IgM, washed, and then incubated at 37°C for the indicated time points. At each time point the cells were fixed, and surface BCR expression was monitored by FACS using streptavidin conjugated to Cy5. As a control, a directly Cy5-conjugated Fab was used to monitor initial surface BCR levels. The mean fluorescence intensity (MFI) for the Cy5-conjugated Fab did not change significantly during the course of the assay, suggesting that the Fab remained associated with IgM and was internalized (data not shown). However, we do see a decrease in the presence of the biotinylated Fab on the B cell surface (Fig. 5A). In the GFP- and c-Cbl-transfected cells, the biotinylated Fab equilibrates with the BCR intracellular pool in the first hour of the assay and establishes a steady-state level (Fig. 5A). In contrast, Bal-17 expressing SLAP and c-Cbl had a steady decline in surface BCR levels (Fig. 5A). This decline in surface BCR levels required enzymatically active c-Cbl as coexpression of a mutant of c-Cbl lacking ubiquitin ligase activity (C3AHN) had no effect on BCR levels (Fig. 5A). Expression of Cbl-b with SLAP had no effect on BCR levels, suggesting that c-Cbl and SLAP cooperate in down-modulation of the BCR complex (Fig. 5A). Alterations in BCR recycling also required an intact SH2 and C-terminal domain of SLAP, whereas the SH3 domain was not essential for alterations in BCR recycling (Fig. 5B). Last, the decline in BCR levels upon SLAP and c-Cbl expression depended on Src family kinase activity because treatment with PP2, a known Src family kinase inhibitor, prevented further decline in BCR levels after initial equilibration (Fig. 5C). These studies demonstrate that SLAP and c-Cbl function in combination to alter the dynamics of constitutive BCR recycling leading to decreased surface BCR levels. (Fig. 5D)
Fig. 5.
Altered BCR recycling upon SLAP and c-Cbl expression in BAL-17. (A) Ligand-independent recycling of internalized IgM. BAL-17 cells were transiently transfected with GFP and c-Cbl, SLAP-GFP and c-Cbl, SLAP-GFP and Cbl-b, or SLAP-GFP and a E3 ubiquitin ligase-deficient mutant of c-Cbl(C3AHN). Four hours after transfection, surface IgM was labeled with a biotinylated F(ab) specific for IgM. After washing, the cells were incubated at 37°C for 0, 1, 3, or 4 h and immediately fixed in 1.6% paraformaldehyde, and the remaining surface IgM was detected by using streptavidin-Cy 5. The data are expressed as the average of three independent transfections (±SEM). (B) BAL-17 cells were transiently transfected with GFP-SLAP alone, GFP-SLAP(SH2*) and c-Cbl, GFP-SLAP(SH3*) and c-Cbl, or GFP-SLAP(DC) and c-Cbl. The experiment was performed as described in A. The data are expressed as the average of three independent transfections (±SEM). (C) BAL-17 cells were transiently transfected with GFP-SLAP and c-Cbl, and the experiment was performed as described in A, except that at the 1-h time point, the cells were split and the Src family kinase inhibitor PP2 was added to one-half of the cells. The data are expressed as the average of three independent transfections (±SEM). (D) Model of how SLAP as an adaptor of c-Cbl may lead to (1) increased internalization, (2) decreased recycling, and (3) ubiquitination of activated components of the BCR complex targeting them for degradation in the lysozome or proteosome.
Discussion
The data presented in this article suggest that SLAP and c-Cbl function through intersecting pathways as evidenced by the alterations seen in B cell development in the SLAP and c-Cbl doubly deficient mice. In addition, our data suggests a model whereby SLAP and c-Cbl in combination alter BCR recycling, which leads to decreased surface and total BCR levels. SLAP, as an adaptor of c-Cbl, may lead to (1) increased internalization, (2) decreased recycling, and (3) ubiquitination of activated components of the BCR complex targeting them for degradation in the lysosome or proteosome (Fig. 5D). These alterations in BCR recycling require Src family kinase activity, an intact SH2 and C-terminal domain of SLAP and enzymatically active c-Cbl.
Together, SLAP and c-Cbl could alter the dynamics of BCR recycling through at least two mechanisms. First, SLAP and c-Cbl could simply link the BCR to clathrin-associated endocytic machinery and thus increase the rate of receptor endocytosis. Internalization of the BCR complex is primarily a clathrin-dependent process, with ≈70% of receptor internalized through this pathway (23). In B cells, clathrin is associated with lipid rafts and is phosphorylated by Src family kinases residing in the rafts after BCR cross-linking (24). Phosphorylation of clathrin leads to its assembly and association with the adaptor related protein complex (AP2) resulting in BCR internalization (24). The AP2 complex also can associate with c-Cbl through its association with CIN85. CIN85 has been shown to associate with endophilin, an endocytic molecule that has been proposed to induce negative membrane curvature and assist in endosome formation (25). Second, SLAP may function to adapt the E3 ubiquitin ligase, c-Cbl, to the BCR complex, facilitating the ubiquitination of components of the BCR complex. A number of signaling proteins downstream of the BCR previously have been shown to be ubiquitinated by c-Cbl, including Lyn and Syk (17, 26, 27). Previous studies have not addressed whether other components of the BCR complex are ubiquitinated. However, it is possible that ubiquitination of components of the BCR complex may lead to their internalization and /or retention in the sorting endosome as seen with growth factor receptors (28). Internalized components of the BCR complex then may be shuttled to the late endosome and lysosomes for degradation. Our observation that the E3 ubiquitin ligase activity of c-Cbl is required for SLAP and c-Cbl-dependent BCR down-modulation provides strong evidence for the aforementioned mechanism.
Internalization and recycling of the BCR is a complex process that can be regulated in a number of different ways. Studies in B cell lines have shown that the cytoplasmic chain of Igα is required for recruitment of the BCR complex to clathrin-coated pits as well as constitutive BCR complex internalization (29). Furthermore, tyrosine phosphorylation of the first tyrosine in the immunoreceptor tyrosine activation motif (ITAM) of Igα is required for constitutive BCR internalization (29). Studies using Igα and Igβ containing platelet-derived growth factor receptor (PDGFR) chimeric receptors have indicated that the ITAMs of Igα are required for constitutive internalization of the BCR complex (30). These studies also suggested a role for Igβ in trafficking the BCR complex to an MHC class II-rich late endosomal compartment (30). Mice with mutations in Igα have alterations in BCR levels on developing B cells (31, 32). B cells developing in mice in which the Igα ITAM tyrosines were replaced with phenylalanines displayed increased BCR levels on most B cell subsets (32). In contrast, developing B cells expressing a BCR complex with two intracellular Igα signaling chains have decreased BCR levels, increased constitutive BCR internalization, and an anergic phenotype (31). Together, these studies provide strong evidence that tyrosine phosphorylation of both Igα and Igβ regulates intracellular trafficking of the BCR complex. How SLAP is involved in this process is only beginning to be elucidated.
Developing B and T lymphocytes both express SLAP and appear to have a conserved mechanism for regulating antigen-receptor levels. Our studies suggest that SLAP functions as an adaptor of the E3 ubiquitin ligase c-Cbl, thereby controlling antigen-receptor expression levels by targeting components of the antigen receptor for degradation. Regulation of antigen-receptor levels during lymphocyte development may act to dampen ligand-dependent and ligand-independent signals from the antigen-receptor complex, possibly allowing the selection of a broader range of antigen receptors with greater diversity of receptor affinity. Diversity of antigen-receptor specificity is essential for generation of a dynamic and plastic immune response to pathogens. Thus, SLAP may be necessary to generate an optimal lymphocyte repertoire.
Materials and Methods
Mice.
The SLAP-deficient, c-Cbl-deficient, and SLAP c-Cbl doubly deficient mice have been described previously (8, 14, 16). The mice were backcrossed to C57BL/6 (B6) for at least five generations. All mice were maintained under specific pathogen-free conditions and were analyzed between 8 and 12 weeks of age for all studies performed. The University of California, San Francisco, Animal Use and Care Committee approved all of the animal research methods used in this study.
Cell Lines and Cell Culture.
The mature mouse B cell lymphoma Bal-17 was used for all studies described. Bal-17 B cells were cultured in RPMI medium 1640 (GIBCO BRL, Carlsbad, CA) supplemented with 10% FCS, 2 mM l-glutamine (GIBCO BRL), 100 international units/ml penicillin/streptomycin (GIBCO BRL), nonessential amino acids, (GIBCO BRL), sodium pyruvate (GIBCO BRL), and 100 μM 2-mercaptoethanol (Sigma, St. Louis, MO).
Flow Cytometry.
Single-cell suspensions from bone marrow, spleen, and peritoneal lavage were stained in FACS buffer [PBS supplemented with 2% FCS, 2 mM l-glutamine (GIBCO BRL), 100 international units/ml penicillin/streptomycin (GIBCO BRL), and 2 mM EDTA] with monoclonal antibodies conjugated with fluorescein isothiocyanate (FITC), phycoerythrin (PE), TRI-COLOR (TC), PerCP (PCP), PerCP-Cy 5.5, or allophycocyanin (APC) as previously described (8, 11). The monoclonal antibodies used were anti-CD45R (B220), anti-IgD, anti-IgM, anti-CD19, anti-CD21, and anti-CD23 (BD PharMingen or eBioscience, both in San Diego, CA). Data were collected on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) system and analyzed by using FlowJo software (Tree Star, Ashland, OR). Data are presented as the mean ± SEM, and statistical analysis was performed by using a two-tailed Student's t test with significance set at P < 0.05.
Transfected BAL-17 B cells were stained in FACS buffer with a goat anti-mouse F(ab′)2 (10 μg/ml) (Jackson Immunoresearch, West Grove, PA) conjugated with biotin. Biotinylated monoclonal antibodies were visualized with streptavidin PerCP (Caltag, Carlsbad, CA). Data were collected and analyzed as described above.
Expression Constructs.
Construction of plasmids encoding GFP-tagged SLAP were described previously (16). Xpress-tagged c-Cbl and Cbl-b constructs were a gift from Y.-C. Liu (La Jolla Institute for Allergy and Immunology, San Diego, CA). Hemagglutinin-tagged c-Cbl construct with mutations in the ring finger that abolish its E3 ligase activity was constructed as previously described (16, 33).
Transient Cell Transfection.
BAL-17 B cells (20 × 106) were transfected by electroporation (300 V, 960 μF) with 40 μg of total plasmid DNA as previously described (34). Transfections were performed either 4 or 18 h before examination.
BCR Stimulation.
BAL-17 B cells were removed from culture, washed, and rested in PBS containing Mg2+ and Ca2+ for 30 min at 37°C. Cells then were stimulated with a goat anti-mouse IgM F(ab′)2 (Jackson Immunoresearch) at 10 μg/ml final concentration for 5 min and then lysed in 1% Nonidet P-40 (Calbiochem, San Diego, CA) lysis buffer plus inhibitors and proteins solubilized as previously described (34).
GST Pull-Down Assay.
Expression of GST and GST SLAP was induced in the bacterial host BL-21, and proteins were purified and coupled to GST agarose as previously described (5). Lysates from BAL-17 B cells stimulated for 0, 5, 10, and 20 min with F(ab′)2 were prepared as described above. The lysates were precleared with GST containing glutathione-agarose beads three times for 30 min at 4°C on a rotator. SLAP-GST-coupled beads were added to the precleared lysates and rotated/tumbled for 2 h at 4°C. After tumbling, the samples were washed four times in lysis buffer, boiled for 3 min in SDS sample buffer, and run on a SDS/polyacrylamide gel. Proteins were transferred onto Immobilon-P membranes (Millipore, Billerica, MA), blocked in TBST/BSA (50 mM Tris, pH 7.4/150 mM NaCl/0.1% Tween 20/3% BSA) for 2 h at room temperature and incubated overnight in the same buffer supplemented with either the mouse anti-phosphotyrosine (4G10; Cell Signaling Technology, Danvers, MA) (8, 35). The blotted proteins were visualized by using horseradish peroxidase-conjugated secondary antibody (Jackson Immunoresearch) and detected by using enhanced chemiluminescence (PerkinElmer, Wellesley, MA).
Coimmunoprecipation.
Bal-17 (20 × 106) stably expressing either SLAP-GFP or GFP were stimulated and lysed as outlined above. Lysates were immunoprecipitated for 90 min with a rat anti-GFP antibody cross-linked to agarose beads (RQ2; MBL International, Woburn, MA). Immunoprecipitates were washed four times with ice-cold lysis buffer and resuspended in SDS/PAGE loading buffer. Samples were separated on a 12% acrylamide gel, and Western blotting analyses were performed as outlined above. The blotted proteins were visualized by using a mouse anti-phosphotyrosine antibody (4G10; Cell Signaling Technology), rabbit anti-Igα (PR-α; a gift from J. Cambier, University of Colorado Health Sciences Center and National Jewish Medical and Research Center, Denver, CO) or rabbit anti-GFP (A6455; Molecular Probes, Carlsbad, CA) as described above.
Immunofluorescence.
Transfected cells were washed and allowed to settle onto poly(l-lysine)-coated plates. Samples were fixed in fixation medium (Caltag) for 20 min at room temperature, washed, and permeabilized with permeabilization medium (Caltag) for 10 min at room temperature. Cells were washed and incubated in blocking buffer [0.2% BSA/10% normal goat serum (NGS)/1% 24G2 (Fc Block) in PBS] for 10 min at room temperature, followed by incubation with a rabbit heterosera specific for c-Cbl (C-15; Santa Cruz Biotechnology, Santa Cruz, CA) for 60 min at 37°C or overnight at 4°C. After washing in blocking buffer, samples again were incubated in blocking buffer, and then the secondary antibody (Alexa 555 goat anti-rabbit IgG; Molecular Probes) was added for 20 min at room temperature. Samples were washed, mounted in gel/mount (Biomeda, Foster City, CA), and visualized, and images were captured as previously described (9).
BCR Recycling Assay.
Transfected BAL-17 cells were washed and placed on ice for 30 min in RPMI medium 1640 containing biotinylated goat anti-mouse IgM F(ab′)2 (10 μg/ml) (Jackson Immunoresearch). The cells were washed twice and placed in warm growth media, and cells were removed at time 0, 1, 3, and 4 h after return to culture. Once the cells were removed, they immediately were fixed in 1.6% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in FACS buffer until the end of the assay. After fixation, cells were washed in FACS buffer and stained for 10 min with allophycocyanin-conjugated streptavidin (Caltag). The cells were washed with FACS buffer twice and either they were maintained in 1.6% paraformaldehyde for later analysis or the data were immediately collected on a FACSCalibur system and analyzed by using FlowJo software. GFP+ cells were analyzed for surface IgM staining, and the average geometric mean of fluorescence intensity (GeoMFI) ± the SEM for three independent transfections per group was determined. As a control for Fab binding, a separate aliquot of transfected cells was stained with the same Fab used above but directly conjugated to Cy-5 (Jackson Immunoresearch). The cells were treated and analyzed as described above.
Acknowledgments
We thank Yun-Cai Liu for providing the Xpress-tagged c-Cbl and Cbl-b constructs and John Cambier for providing the PR-α anti-Ig-α antisera. We also thank all of the members of the Weiss Laboratory for their helpful comments and suggestions and Virginia D. Winn and Susan Levin for their critical review of the manuscript. This work was supported by National Institutes of Health Grants 5K08AI055873-04 (to L.L.D.) and CA72531 (to A.W.).
Abbreviations
- SLAP
Src-like adaptor protein
- TCR
T cell receptor
- BCR
B cell receptor
- SH
Src homology
- DP
double-positive.
Footnotes
The authors declare no conflict of interest.
References
- 1.Goodnow CC, Glynne R, Akkaraju S, Rayner J, Mack D, Healy JI, Chaudhry S, Miosge L, Wilson L, Papathanasiou P, Loy A. Adv Exp Med Biol. 2001;490:33–40. doi: 10.1007/978-1-4615-1243-1_4. [DOI] [PubMed] [Google Scholar]
- 2.Goodnow CC. Lancet. 2001;357:2115–2121. doi: 10.1016/s0140-6736(00)05185-0. [DOI] [PubMed] [Google Scholar]
- 3.Tang J, Sawasdikosol S, Chang JH, Burakoff SJ. Proc Natl Acad Sci USA. 1999;96:9775–9780. doi: 10.1073/pnas.96.17.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey A, Duan H, Dixit VM. J Biol Chem. 1995;270:19201–19204. doi: 10.1074/jbc.270.33.19201. [DOI] [PubMed] [Google Scholar]
- 5.Sosinowski T, Pandey A, Dixit VM, Weiss A. J Exp Med. 2000;191:463–474. doi: 10.1084/jem.191.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roche S, Alonso G, Kazlauskas A, Dixit VM, Courtneidge SA, Pandey A. Curr Biol. 1998;8:975–978. doi: 10.1016/s0960-9822(98)70400-2. [DOI] [PubMed] [Google Scholar]
- 7.Manes G, Bello P, Roche S. Mol Cell Biol. 2000;20:3396–3406. doi: 10.1128/mcb.20.10.3396-3406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sosinowski T, Killeen N, Weiss A. Immunity. 2001;15:457–466. doi: 10.1016/s1074-7613(01)00195-9. [DOI] [PubMed] [Google Scholar]
- 9.Myers MD, Dragone LL, Weiss A. J Cell Biol. 2005;170:285–294. doi: 10.1083/jcb.200501164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldhahn N, Schwering I, Lee S, Wartenberg M, Klein F, Wang H, Zhou G, Wang SM, Rowley JD, Hescheler J, et al. J Exp Med. 2002;196:1291–1305. doi: 10.1084/jem.20020881. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Dragone LL, Myers MD, White C, Sosinowski T, Weiss A. J Immunol. 2006;176:335–345. doi: 10.4049/jimmunol.176.1.335. [DOI] [PubMed] [Google Scholar]
- 12.Glynne R, Ghandour G, Rayner J, Mack DH, Goodnow CC. Immunol Rev. 2000;176:216–246. doi: 10.1034/j.1600-065x.2000.00614.x. [DOI] [PubMed] [Google Scholar]
- 13.Loreto MP, Berry DM, McGlade CJ. Mol Cell Biol. 2002;22:4241–4255. doi: 10.1128/MCB.22.12.4241-4255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naramura M, Kole HK, Hu RJ, Gu H. Proc Natl Acad Sci USA. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, Langdon WY, Bowtell DD. Mol Cell Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers MD, Sosinowski T, Dragone LL, White C, Band H, Gu H, Weiss A. Nat Immunol. 2006;7:57–66. doi: 10.1038/ni1291. [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Yang C, Elly C, Liu YC. J Biol Chem. 2004;279:43646–43653. doi: 10.1074/jbc.M404082200. [DOI] [PubMed] [Google Scholar]
- 18.Thien CB, Langdon WY. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 19.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 20.de Melker AA, van der Horst G, Calafat J, Jansen H, Borst J. J Cell Sci. 2001;114:2167–2178. doi: 10.1242/jcs.114.11.2167. [DOI] [PubMed] [Google Scholar]
- 21.Donovan JA, Wange RL, Langdon WY, Samelson LE. J Biol Chem. 1994;269:22921–22924. [PubMed] [Google Scholar]
- 22.Rao N, Lupher ML, Jr, Ota S, Reedquist KA, Druker BJ, Band H. J Immunol. 2000;164:4616–4626. doi: 10.4049/jimmunol.164.9.4616. [DOI] [PubMed] [Google Scholar]
- 23.Stoddart A, Jackson AP, Brodsky FM. Mol Biol Cell. 2005;16:2339–2348. doi: 10.1091/mbc.E05-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoddart A, Dykstra ML, Brown BK, Song W, Pierce SK, Brodsky FM. Immunity. 2002;17:451–462. doi: 10.1016/s1074-7613(02)00416-8. [DOI] [PubMed] [Google Scholar]
- 25.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. Nature. 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 26.Yankee TM, Keshvara LM, Sawasdikosol S, Harrison ML, Geahlen RL. J Immunol. 1999;163:5827–5835. [PubMed] [Google Scholar]
- 27.Rao N, Ghosh AK, Ota S, Zhou P, Reddi AL, Hakezi K, Druker BK, Wu J, Band H. EMBO J. 2001;20:7085–7095. doi: 10.1093/emboj/20.24.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane-Robinson K, et al. J Biol Chem. 2003;278:28950–28960. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- 29.Cassard S, Salamero J, Hanau D, Spehner D, Davoust J, Fridman WH, Bonnerot C. J Immunol. 1998;160:1767–1773. [PubMed] [Google Scholar]
- 30.Siemasko K, Eisfelder BJ, Stebbins C, Kabak S, Sant AJ, Song W, Clark MR. J Immunol. 1999;162:6518–6525. [PubMed] [Google Scholar]
- 31.Reichlin A, Gazumyan A, Nagaoka H, Kirsch KH, Kraus M, Rajewsky K, Nussenzweig MC. J Exp Med. 2004;199:855–865. doi: 10.1084/jem.20031140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraus M, Pao LI, Reichlin A, Hu Y, Canono B, Cambier JC, Nussenzweig MC, Rajewsky K. J Exp Med. 2001;194:455–469. doi: 10.1084/jem.194.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ota S, Hazeki K, Rao N, Lupher ML, Jr, Andoniou CE, Druker B, Band H. J Biol Chem. 2000;275:414–422. doi: 10.1074/jbc.275.1.414. [DOI] [PubMed] [Google Scholar]
- 34.Gupta N, DeFranco AL. Mol Biol Cell. 2003;14:432–444. doi: 10.1091/mbc.02-05-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell KS, Hager EJ, Friedrich RJ, Cambier JC. Proc Natl Acad Sci USA. 1991;88:3982–3986. doi: 10.1073/pnas.88.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]