Abstract
An important role of IgG antibodies in the defense against microbial infections is to promote the ingestion and killing of microbes by phagocytes. Here, we developed in vivo and in vitro approaches to ask whether opsonization of particles with IgG enhances intracellular targeting of lysosomes to phagosomes. To eliminate the effect of IgG on the ingestion process, cells were exposed to latex beads at 15–20°C, which allows engulfment of both IgG-coated and uncoated beads but prevents the fusion of lysosomes with phagosomes. Upon shifting the temperature to 37°C, phagosomes containing IgG beads matured significantly faster into phagolysosomes as judged by colocalization with lysosomal markers. The IgG effect was independent of other particle-associated antigens or serum factors. Lysosome/phagosome attachment was also quantified biochemically with a cytosol-dependent scintillation proximity assay. Interactions were enhanced significantly in reactions containing cytosol from mouse macrophages that had been exposed to IgG-coated beads, indicating that IgG signaling modulates the cytosolic-targeting machinery. Similar results were obtained with cytosol from primary human monocytes, human U-937 histiocytic lymphoma cells and from Chinese hamster ovary (CHO) cells transfected with a human IgG (Fcγ) receptor. IgG-induced activation is shown to affect the actin-dependent tethering/docking stage of the targeting process and to proceed through a pathway involving protein kinase C. These results provide a rare example of an extracellular signal controlling membrane targeting on the level of tethering and docking. We propose that this pathway contributes to the role of antibodies in the protection against microbial infections.
Keywords: membrane fusion, protein kinase C, scintillation proximity assay, Fc receptor, macrophages
Antibodies of the IgG class constitute a critical part of the humoral immune system (1). Complete absence of immunoglobulins causes high susceptibility to infections by microorganisms that is managed effectively by treatment with IgG (2, 3). The protective effect of IgG results in part from its stimulatory effects on professional phagocytes such as monocytes, macrophages, and neutrophils. During phagocytosis, microbes or other particles are enveloped in a patch of membrane and stored in intracellular vacuoles termed phagosomes. Phagosomes subsequently mature into phagolysosomes through fusion with endosomes and lysosomes that deliver proteins involved in luminal acidification, killing, and degradation (4).
Although phagocytes efficiently internalize even unmodified particles such as latex beads (5), opsonization of substrates with IgG triggers signaling cascades, through clustering of cell surface Fcγ receptors, that accelerate the engulfment process and activate the generation of microbicidal oxygen and nitrogen species (6–9).
Studies with intracellular pathogens have suggested that IgG signaling extends beyond the initial engulfment phase. Mycobacterium tuberculosis and Toxoplasma gondii, for example, enter cells by phagocytosis and then replicate intracellularly in part by preventing the fusion of microbe-bearing phagosomes with lysosomes (10, 11). By contrast, in macrophages containing dead microbes or live microbes opsonized with IgG, lysosome/phagosome fusion proceeds and the pathogens are destroyed more effectively (11–14). However, whether IgG signaling promotes lysosome/phagosome targeting only indirectly through enhanced uptake and killing, or also directly by modulating the maturation of formed phagosomes, has not yet been determined.
Here, we developed in vivo and in vitro approaches that allowed us to specifically focus on the lysosome/phagosome targeting process. Our results indicate that IgG signaling promotes lysosome/phagosome targeting on the level of tethering or docking.
Results
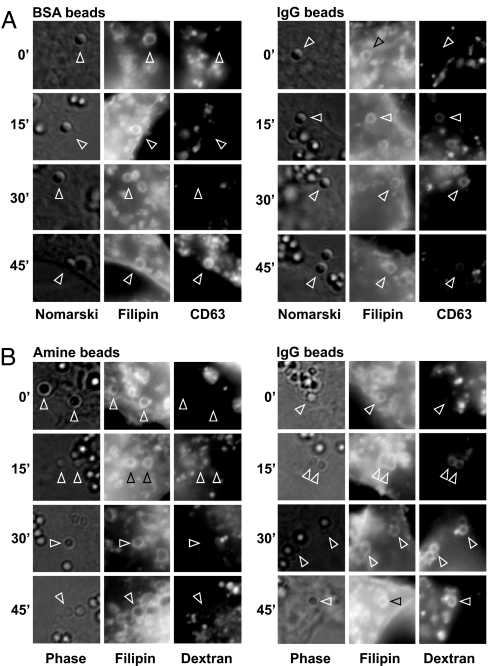
In Fig. 1, we asked whether opsonization of particles with IgG alone can influence the intracellular fusion of lysosomes with phagosomes. RAW 264.7 mouse macrophages were infected for 2 days with a lentivirus expressing the late endosomal/lysosomal marker CD63 fused to cherry fluorescent protein. The cells were then switched to serum-free medium and exposed to 1-μm latex beads that had been cross-linked to BSA or IgG. To eliminate the effects of IgG on the kinetics of the engulfment process, beads were added to macrophages for 60 min at 15°C, washed, and incubated at 15°C for an additional 60 min. At 15°C, beads are taken up as indicated by staining of phagosomes with the membrane dye filipin; however, the phagosomes fail to fuse with CD63-marked vesicles (Fig. 1A, 0-min chase). When cells were subsequently shifted to 37°C, fusion resumed, and CD63 staining around BSA beads became visible after ≈45 min (Fig. 1A and Table 1). By contrast, when IgG-covered beads were used, CD63 staining around the majority of phagosomes became evident as early as 15 min. Similar results were obtained when lysosomes were labeled with fluorescent dextran (Fig. 1B). These results support the conclusion that opsonization of particles with IgG is sufficient to accelerate lysosome/phagosome targeting.
Fig. 1.
IgG opsonization promotes phagosome maturation in vivo. (A) On day 0, RAW 264.7 cells were set up on coverslips in a 24-well plate. On day 1, the medium was replaced with 0.3 ml of culture supernatant containing lentivirus encoding CD63 fused to cherry fluorescent protein. On day 2, cells were refed fresh medium and grown for another 2 days. Cultures were switched to suspensions of medium B plus 1-μm amine latex beads coupled to BSA or to mouse IgG as indicated. Plates were centrifuged at 1,000 × g for 2 min and incubated at 15°C for 1 h, washed three times with PBS to remove unattached beads, and incubated again for 1 h at 15°C. The plates were then moved to 37°C for the indicated time. Cells were fixed, stained with filipin (15), and analyzed by microscopy. (B) RAW cells were set up as above and on day 1 received medium C plus 0.1 mg/ml fixable tetramethylrhodamine-conjugated dextran (10 kDa; Invitrogen). After incubation at 37°C overnight, the cells were switched to medium A and exposed to 1-μm amine beads or to amine beads conjugated to mouse IgG. The protocol for bead uptake and chase was as in A. Arrowheads point at selected phagosomes.
Table 1.
Fusion of lysosomes with phagosomes containing BSA- versus IgG-coated latex beads
| Opsonin | Length of 37°C chase, min | Phagosomes | Phagolysosomes | % fused |
|---|---|---|---|---|
| BSA | 0 | 165 | 2 | 1.2 |
| 15 | 138 | 3 | 2.2 | |
| 30 | 283 | 23 | 8.1 | |
| 45 | 290 | 233 | 80.3 | |
| IgG | 0 | 174 | 1 | 0.6 |
| 15 | 396 | 368 | 92.9 | |
| 30 | 447 | 413 | 92.4 | |
| 45 | 258 | 243 | 94.2 |
RAW 264.7 macrophages were infected with lentivirus expressing ChFP-CD63 and exposed to 1-μm latex beads coated with BSA or IgG as described in the Fig. 1A legend. Phagosomes were identified based on positive filipin staining. Phagolysosomes were defined as filipin-positive phagosomes surrounded by rings of CD63 staining. Data are from two independent experiments.
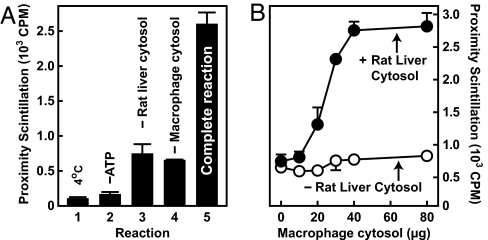
As an approach to exploring the biochemical basis for this activation, lysosome/phagosome interactions were studied with a cell-free scintillation proximity assay that has been described previously (15). The key components of this assay are phagosomes containing scintillant beads and lysosomes labeled with tritiated cholesteryl ether. Because of the short range of tritium-derived β particles and as a consequence of geometric constraints, scintillation requires immediate contact between lysosomes and phagosomes (15–17). Using this assay, we previously showed that lysosomes and phagosomes will attach when reactions are supplemented with ATP and rat liver cytosol (15). In these experiments, lysosomes were added as a component of postnuclear supernatants that contain both membranes and low-salt macrophage cytosol. In Fig. 2, we sought to determine whether proteins from macrophage cytosol also are required for the in vitro reaction. When postnuclear supernatants were centrifuged and lysosomes supplied as part of the resultant pellet, scintillation remained low, unless reactions were resupplied with cytosol from unlabeled macrophages (Fig. 2 A and B). These data show that an activity from macrophage cytosol is required for lysosomes and phagosomes to attach in the cell-free assay.
Fig. 2.
Lysosome/phagosome interactions in vitro require macrophage cytosol. (A) Lysosome/phagosome targeting was measured with a cell-free scintillation proximity assay. Complete reactions contained 3H-cholesteryl oleoyl ether-labeled lysosomes, J774 cell phagosomes containing scintillant latex beads plus or minus cytosol from J774 cells and rat liver, and an ATP-regenerating system. After 1 h of incubation, proximity scintillation was measured and is expressed in cpm. (B) Reactions were as in A except that macrophage cytosol concentrations were varied in the absence and presence of 6 mg/ml rat liver cytosol.
Why cytosol from both rat liver and macrophages is required during the in vitro assay is currently unclear. The factor from macrophages might not be present in rat liver or it might be sensitive to increased salt concentrations. In support of the latter hypothesis, preparation of macrophage cytosol with the isotonic buffer used for liver homogenization had less activity (V.T. and A.N., unpublished data).
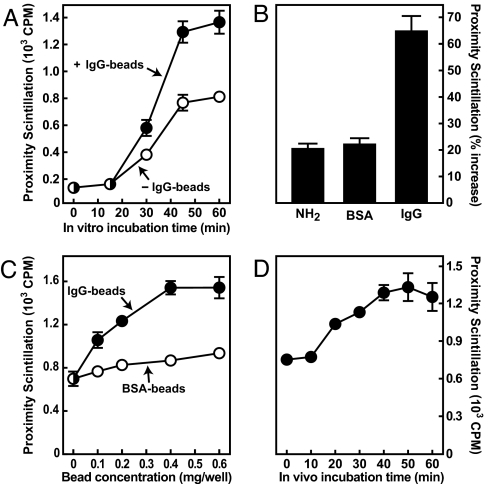
Using the cell-free assay, we next asked whether the IgG-induced acceleration of lysosome/phagosome targeting that was observed in intact cells might involve the activation of cytosolic factors. Scintillation proximity reactions were set up with cytosol extracted from J774 macrophages that had been cultured for 1 h in the absence or presence of IgG beads. Analogous to the results obtained by microscopy, cytosol from both cell populations promoted lysosome/phagosome interactions, but cytosol from cells exposed to IgG beads was significantly more potent (Fig. 3A). Optimal activation required the presence of IgG on the particles as amine-coated beads or beads that had been coupled to BSA were less effective (Fig. 3 B and C). Activation of macrophage cytosol by IgG beads became detectable as early as 20 min after addition of beads (Fig. 3D). Direct exposure of unconditioned macrophage cytosol to IgG beads before in vitro reactions had no stimulating effect (data not shown).
Fig. 3.
IgG-opsonized beads activate a cytosolic factor required for promoting lysosome/phagosome interactions. Cell-free assays were performed as in Fig. 2A. (A) Low-salt cytosol was from J774 cells grown for 1 h in the absence or presence IgG-coated beads in medium B. (B) Low-salt cytosol was from J774 cells grown in medium B for 1 h in the absence of beads, with 0.25 mg per well of 6-μm amine beads (NH2), amine beads coupled to BSA, or amine beads coupled to mouse IgG (data were calculated by comparing the effects of low-salt cytosol from bead-treated versus untreated controls). (C) Low-salt cytosol was from J774 cells exposed to the indicated concentrations of 6-μm BSA beads (open circles) or IgG beads (filled circles) in medium B for 1 h. (D) Low-salt cytosol was from J774 cells exposed to 0.2 mg/ml 6-μm IgG beads in medium B for the indicated time. Background scintillation counts in reactions performed without macrophage cytosol were 148 ± 15 cpm (A), 176 ± 11 cpm (B), 446 cpm (C), and 195 ± 41 cpm (D).
As expected from the microscopy data, activation of cytosol by IgG beads also was seen in RAW 264.7 macrophages as well as in primary human monocytes, human U-937 histiocytic lymphoma cells, and primary mouse peritoneal macrophages (Table 2). Together, the above data show that exposure of macrophages to IgG-opsonized particles leads to the activation of one or more cytosolic factors involved in lysosome/phagosome targeting.
Table 2.
Effect of IgG-conjugated beads on different cell types
| Activity, cpm | ||||
|---|---|---|---|---|
| Cell line | −IgG beads | +IgG beads | % increase | P value |
| HEK 293* | 732 ± 89 | 778 ± 41 | 6.2 | 0.61 |
| ARPE-19* | 716 ± 46 | 792 ± 94 | 10.6 | 0.45 |
| GC-1 spg* | 840 ± 18 | 970 ± 94 | 15.5 | 0.13 |
| NB4* | 1,073 ± 75 | 1,514 ± 180 | 41.2 | 0.093 |
| HL-60* | 744 ± 29 | 661 ± 17 | −11.2 | 0.037 |
| Primary human monocytes† | 870 ± 47 | 1,384 ± 90 | 59.0 | 0.016 |
| RAW 264.7* | 956 ± 92 | 1,314 ± 67 | 37.5 | 0.013 |
| J774* | 637 ± 28 | 1,438 ± 107 | 125.7 | 0.0074 |
| U-937* | 869 ± 58 | 993 ± 42 | 14.3 | 0.0071 |
| Primary mouse peritoneal macrophages* | 702 ± 78 | 997 ± 53 | 42.0 | 0.0035 |
Lysosome/phagosome targeting was measured by proximity scintillation assay as in Fig. 2A. Low-salt cytosol was from the indicated cell types grown in the absence or presence of 0.25 mg/ml 6-μm IgG beads in medium B for 1 h. Activity data indicate mean ± SD (n = 3). For each cell line, P values were calculated by paired, two-tailed Student's t test by comparing data sets obtained with unconditioned (−IgG beads) and IgG bead-conditioned (+IgG beads) cytosol. HEK 293, immortalized human embryonic kidney epithelial cells (34); ARPE-19, human retinal pigment epithelial cells (35); GC-1 spg, immortalized mouse spermatogonia (36); NB4, human promyelocytic leukemia cells (37); HL-60, human promyelocytic leukemia cells (38); RAW 264.7, immortalized mouse macrophages (39); J774, mouse macrophage tumor cell line (40); U-937, human histiocytic lymphoma cells (41).
*Background activity in the absence of low-salt cytosol, 272 ± 19 cpm.
†Data for human monocytes are from a separate experiment. Background activity in the absence of low-salt cytosol, 264 ± 52 cpm.
Next, we sought to further characterize this signaling pathway. IgG is recognized by a family of plasma membrane proteins called Fcγ receptors (18). Particle-induced clustering of Fcγ receptors can trigger signaling cascades whose downstream targets include serine/threonine kinases of the protein kinase C (PKC) family (19, 20). Conventional PKCs have been found on pinched-off phagosomes (21–24), and their concentration has been reported to be ≈35 times higher around IgG-opsonized versus BSA-coated phagosomes (22). In addition, a PKC agonist, phorbol 12-myristate 13-acetate (PMA), has been reported to promote phagosome maturation (25).
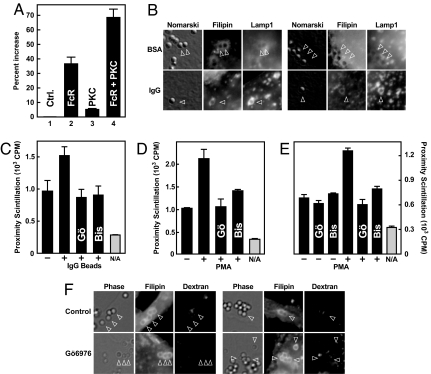
To test whether Fcγ receptors and PKC can cooperate in relaying an IgG signal to the lysosome/phagosome targeting machinery, cDNAs expressing human Fcγ receptor IIa (FcγRIIa) and prototypical PKC-α were transfected into CHO cells. Transfection of FcγRIIa renders CHO cells phagocytic and capable of phagolysosome formation (26). Cells were transfected for 20 h and incubated in the absence or presence of IgG beads. Cytosol extracts were then tested in the cell-free scintillation proximity assay. In mock-transfected CHO cells, there was no change after exposure to IgG beads (Fig. 4A, set 1). In cells transfected with FcγRIIa, however, activity increased by 37% (Fig. 4A, set 2). Only a small effect was seen after transfection of PKC-α alone (Fig. 4A, set 3), but a 68% increase in response to IgG resulted when cells were transfected with both PKC-α and FcγRIIa (Fig. 4A, set 4). The effect was blocked when transfected cells were treated with both IgG beads and the PKC-α/β inhibitor Gö6976 (data not shown).
Fig. 4.
Characterization of IgG-induced signaling pathway. (A) CHO-K1 cells were transfected with 0.7 μg each of plasmids expressing human FcγRIIa (pCMV-FcγRIIa-IRES-Neo; FcR) and a yellow fluorescent protein (YFP)/mouse PKC-α fusion protein (pEX-EF1-YFP-PKC-α; PKC). Total DNA amounts were adjusted to 2.1 μg per well by addition of pIRESneo2. Cells were cultured plus or minus human IgG beads in medium A for 1 h at 37°C, harvested, and fractionated. Low-salt cytosol was used for in vitro reactions as in Fig. 2A. Data indicate percentage increase of scintillation in reactions containing low-salt cytosol from IgG-treated versus control cells. (B) CHO-K1 cells were transfected for 48 h with 70 ng each of plasmids expressing FcγRIIa, YFP-PKC-α, and Lamp1-RFP. Cultures were exposed to 1-μm BSA beads or IgG beads at 15°C as in Fig. 1 and chased at 37°C for 15 min. Cells were fixed, stained with filipin, and analyzed by microscopy. For each condition, two representative sets of images are shown. (C) Lysosome/phagosome targeting was measured as in Fig. 2A. Reactions were set up without (gray bars) or with (black bars) low-salt cytosol from J774 cells cultured for 1 h in the absence or presence of 0.2 mg/ml 6-μm IgG beads, 1 μM Gö6976 (Gö), and 10 μM bisindolylmaleimide (Bis). (D) Reactions were set up as in C. Low-salt cytosol was from J774 cells that had been cultured plus or minus 1 μM Gö6976 and 10 μM bisindolylmaleimide for 1 h and then received solvent or 1.62 pM PMA for 1 h. (E) Reactions were set up with low-salt cytosol from J774 cells grown under control conditions. The indicated drugs were added in vitro at the concentrations used above. (F) RAW cells were loaded with fluorescent dextran and exposed to 1-μm IgG beads at 15°C as in Fig. 1B and chased at 37°C for 15 min. Where indicated, 10 μM Gö6976 was present during the final hour at 15°C and during the 37°C chase. For each condition, two representative sets of images are shown. Arrowheads point at selected phagosomes.
To verify these results morphologically, CHO cells were transfected with plasmids expressing FcγRIIa, PKC-α, and the late endosomal/lysosomal membrane protein Lamp1 fused to red fluorescent protein (RFP). The cells then were exposed to BSA-coated or IgG-coated latex beads at 15°C, chased at 37°C, and analyzed by microscopy. After 15 min of chase, Lamp1-RFP staining was strong in IgG bead containing phagosomes but only barely discernable in phagosomes containing BSA beads (Fig. 4B). Analogous to the results in Fig. 1, Lamp1-RFP staining after 45 min was similar for both types of beads, confirming the notion that IgG is not essential for but accelerates phagosome maturation (data not shown).
Next, scintillation proximity assays were performed with low-salt cytosol from J774 cells exposed to IgG beads and the pan-specific PKC inhibitor bisindolylmaleimide or to Gö6976, an inhibitor specific for PKC-α and PKC-β (27). Both drugs abolished the stimulatory effect of IgG beads while leaving the basal activity unaltered (Fig. 4C).
A role for PKCs in regulating lysosomes is supported further by the observation that PMA was able to stimulate lysosome/phagosome attachment when added to cells before cytosol extraction (Fig. 4D) or directly to reactions in vitro (Fig. 4E); in both cases, the effect of PMA was blocked by bisindolylmaleimide and Gö6976.
The effects of PKC inhibition on lysosome/phagosome targeting were also studied by fluorescence microscopy. First, RAW cells were loaded with fluorescent dextran overnight to label lysosomes. The cells then were loaded with IgG beads at 15°C and chased at 37°C in the absence or presence of Gö6976. After 15 min of chase, lysosomes had fused with phagosomes in control cells but not in cells treated with the drug (Fig. 3F). Similar results were obtained when cells were fed BSA beads and treated with PMA (data not shown). Together, the results in Fig. 4 support the conclusion that PKC activity is required for enhanced lysosome/phagosome attachment in response to IgG.
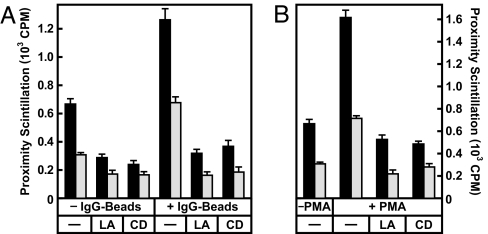
We then asked which stage of the lysosome/phagosome targeting process is stimulated by IgG. Our previous work has shown that lysosome/phagosome fusion is preceded by distinct tethering and docking steps (15). Tethering is a prerequisite for docking and involves the attachment of membranes via filamentous actin (15, 28, 29). To test whether increased lysosome/phagosome targeting in response to IgG requires actin polymerization, cell-free assays were performed with cytosol from J774 cells grown for 1 h in the absence and presence of IgG beads or PMA. Two inhibitors of actin polymerization, latrunculin A and cytochalasin D, were added in vitro. Both drugs prevented lysosome/phagosome attachment in controls as well as in IgG and PMA-conditioned samples (Fig. 5A and B). These data indicate that IgG and PMA modulate the actin-dependent attachment process.
Fig. 5.
IgG signaling activates the tethering/docking phase of lysosome/phagosome targeting. (A) Cell-free scintillation proximity assays were performed as in Fig. 2A. Low-salt cytosol was obtained from J774 cells cultured for 1 h plus or minus IgG beads. In vitro reactions were set up in the absence or presence of 1 μM latrunculin A (LA) and 20 μM cytochalasin D (CD) as indicated and incubated for 1 h at 37°C. Proximity scintillation was determined in a scintillation counter (black bars). Reactions then were supplemented with 0.1 M sodium carbonate (pH 11) and vortexed for 10 s, and scintillation was immediately measured again (gray bars). (B) Cell-free reactions were set up with low-salt cytosol from J774 cells cultured for 1 h in the absence or presence of 1.6 nM PMA as indicated. Latrunculin A and cytochalasin D were added in vitro as in A. Scintillation was measured before and after carbonate treatment as above. Background scintillation counts in the absence of low-salt cytosol were 264 ± 34 cpm (A) and 166 ± 21 cpm (B).
Tethering is a transient process that gives rise to a more long-lived docking stage at which complexes become resistant to actin inhibitors but remain sensitive to disruption by alkaline carbonate (15). If IgG promoted tethering and/or docking, lysosome/phagosome interactions would be expected to increase but drop in response to alkali. This result was in fact observed (Fig. 5A, gray bars), and similar results were obtained in reactions that had been stimulated with PMA (Fig. 5B). From the results in Fig. 5, we conclude that IgG/PKC signaling promotes lysosome/phagosome targeting at the tethering or docking stage.
Discussion
The current results add the stimulation of lysosome/phagosome targeting to the list of effector functions of IgG. Established roles for IgG include the masking of antigenic epitopes, activation of complement, inhibition of B cell activation, and the opsonization of substrates for induction of antibody-dependent cell-mediated cytotoxicity, engulfment by phagocytes, and respiratory burst (30).
Previous work had demonstrated that opsonization of particles with IgG led to an earlier appearance of phagolysosomes, but to what degree this effect results from enhanced killing and engulfment versus activation of the lysosomal targeting machinery had not yet been resolved (11–14). To address this problem, we developed a set of approaches that allowed us to uncouple phagosome formation from the subsequent maturation process. In one set of experiments, we applied a load-and-chase protocol wherein cells were allowed to take up latex beads at 15–20°C and then shifted to 37°C for different periods. The method is based on the observation that at the lower temperatures, both opsonized and unopsonized particles are engulfed but fail to acquire lysosomal markers. Upon a subsequent shift to 37°C, normal trafficking resumes. Fusion of lysosomes with phagosomes containing IgG beads was virtually complete after 15 min but required at least 45 min for unopsonized beads. Similar results were obtained with mouse macrophages and CHO cells transfected with a human Fcγ receptor, strongly arguing that particle-associated IgG, presumably in a complex with cognate receptors, transmits a lysosome-attracting signal across the phagosomal membrane.
To further test this conclusion under more biochemically defined conditions, we used a cell-free scintillation proximity assay in which lysosome/phagosome attachment depends on the supplementation of reactions with macrophage cytosol. In these experiments, interactions were significantly enhanced when cytosol was supplied from macrophages that had been exposed to IgG-coated beads, directly confirming the idea the IgG signaling leads to activation of one or more cytosolic factors.
We reasoned that PKCs were key candidates for components of this signaling pathway. PKCs are induced in response to IgG (19, 20) and are highly enriched on phagosomal membranes surrounding IgG-opsonized versus uncoated particles (22). Several lines of evidence obtained in this study show that PKCs are in fact involved in IgG-induced stimulation of phagolysosome formation: (i) macrophage cytosol could be conditioned with the PKC agonist PMA; (ii) conditioning of macrophage cytosol by IgG beads or PMA was blocked by PKC inhibitors; (iii) transfection of PKC-α synergized with FcγRIIa in rendering CHO cells sensitive to IgG beads as measured in vitro and in vivo; and (iv) at early time points, a PKC-α/β inhibitor blocked the fusion of IgG bead-containing phagosomes with lysosomes.
Known substrates of PKCs include proteins controlling the formation of structures containing filamentous actin that are in turn essential for plasma extrusion during particle engulfment as well as for subsequent fusion of lysosomes with phagosomes (15, 19, 28). Previous work has indicated that during lysosome/phagosome targeting, actin polymerization plays a critical role specifically during the initial tethering stage (15, 29). Our current results support the conclusion that regulation of phagosome maturation is exerted at the early targeting stages, because IgG beads and PMA could enhance lysosome/phagosome attachment in the absence of fusion and in a manner sensitive to actin polymerization inhibitors. Further studies on the macrophage cytosol activity should be useful to extend the understanding of lysosome/phagosome targeting.
Materials and Methods
Reagents.
We obtained BSA from Sigma (St. Louis, MO); bisindolylmaleimide, Gö6976, and PMA from Calbiochem (San Diego, CA); and amine-conjugated latex beads from Polysciences Inc. (Warrington, PA). Latex beads were coupled to BSA or IgG by using glutaraldehyde as described in ref. 31. Other materials were from sources as described in ref. 15. Liposomes were prepared as described in ref. 15 and contained 200 μCi/ml [3H]cholesteryl oleoyl ether (1 Ci = 37 GBq).
Plasmids and Lentivirus.
pLamp1-RFP is described at www.addgene.org/1817 and was kindly provided by Walther Mothes (Yale University School of Medicine, New Haven, CT). pEX-EF1-YFP-PKC-α was generated by the Alliance for Cellular Signaling and was purchased from ATCC (Manassas, VA). A plasmid expressing human FcγRIIA was generated as follows. White blood cells were isolated from human blood (15 ml) by using Ficoll-Paque Plus (Amersham Pharmacia, Piscataway, NJ), and RNA was extracted with Tri reagent (Molecular Research Center, Cincinnati, OH). The FcγRIIa cDNA was amplified by RT-PCR using a Titan One Tube RT-PCR System kit (Roche, Mannheim, Germany) and a pair of primers (5′-TTGAATTCACCATGGAGACCCAAATGTCTC-3′ and 5′-GCGGCCGCTTAGTTATTACTGTTGACATGG-3′). The PCR product was cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) to generate pAC01 and subcloned as an EcoRI–NotI fragment into pIRESneo2 (Clontech, Mountain View, CA) to generate pCMV-FcγRIIa-IRES-Neo. The insert was verified by sequencing. A lentivirus expressing CD63 fused to a cherry-fluorescent protein tag was prepared as described in ref. 32.
Buffers.
ATP regenerating system was prepared as an 8× stock solution containing 2 mM GTP, 8 mM ATP, 40 mM creatine phosphate, and 20 μg/ml creatine kinase, and the pH was set to 7.3 with KOH. Buffer A contains 10 mM KCl, 10 mM Hepes-KOH (pH 7.3), 1.5 mM MgCl2, 1 mM DTT, 1 μg/ml pepstatin, and 5 μg/ml leupeptin. Buffer B contains 100 mM KCl, 40 mM Hepes-KOH (pH 7.3), 3 mM MgCl2, 0.5 mM EGTA, 1 μg/ml pepstatin, and 5 μg/ml leupeptin. Buffer C is buffer B plus 250 mM sucrose. Buffer D is buffer B plus 0.9 M sucrose.
Cell Culture and Transfections.
All cells were grown at 37°C in 8–9% CO2. Medium A is a 1:1 mixture of Ham's F-12 medium and Dulbecco's modified Eagle's medium (DMEM) plus antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin sulfate). Medium B is medium A plus 0.2% (wt/vol) fatty acid-free BSA. Medium C is medium A plus 10% (vol/vol) heat-inactivated FBS. ARPE-19, CHO-K1, J774, and RAW 264.7 cells were grown in medium C. Human embryonic kidney 293 cells were grown in DMEM supplemented with antibiotics and 10% FBS. Primary mouse peritoneal macrophages were obtained from BALB/c mice as described (16). Human monocytes were obtained from the blood (20 ml) of a healthy volunteer by using Ficoll-Paque as described in ref. 33. Mouse peritoneal macrophages, human monocytes, NB4, and U-937 cells were grown in RPMI medium 1640 plus antibiotics and 10% FBS. CHO-K1 cells were transfected by using Fugene 6 reagent (Roche) at a ratio of 3 μl of Fugene 6 per microgram of DNA.
Scintillation Proximity Assay.
Rat liver cytosol and phagosomes containing scintillant beads were prepared as described in ref. 15. Preparation of radioactively labeled lysosomes was performed as follows. J774 cells were set up in a 10-cm dish at 50% confluency in 4 ml of medium C supplemented with 100 μl of 3H-cholesteryl oleoyl ether-containing liposomes. After 16 h, cells were washed three times with PBS and once with buffer A and scraped into 1 ml of buffer A. Cell suspensions were passed eight times through a 22-gauge needle (bent two times) and centrifuged for 5 min at 1,000 × g. Postnuclear supernatants were spun at 16,000 × g for 30 min, and the pellet was resuspended in 1 ml of buffer C. Previously published fractionation experiments have shown that 3H-cholesteryl oleoyl ether and docking activity are confined to the lysosomal fraction (15).
Preparation of Low-Salt Cytosol.
Cells were set up in six-well plates at 70% confluency. The next day, before bead additions, cells were grown for 1 h in medium B. Cells were processed as above without prior labeling. After the final centrifugation step, the supernatant was collected and designated as low-salt cytosol. Further centrifugation at 100,000 × g for 30 min gave results identical to those obtained with 16,000 × g supernatants (V.T. and A.N., unpublished data).
In Vitro Assay.
Reactions were set up, and scintillation was measured as described previously (15). Briefly, reactions (0.4 ml total volume) contained 0.75 mg of phagosomes, J774 membranes containing [3H]cholesteryl oleoyl ether-labeled lysosomes (containing ≈80 μg of protein and ≈40 nCi of tritium), an ATP-regenerating system, 2.4 mg of rat liver cytosol, and, unless otherwise indicated, 40 μg of low-salt macrophage cytosol.
Fluorescence Microscopy and Image Processing.
Cells were fixed with 4% (wt/vol) paraformaldehyde, stained with 50 μg/ml filipin, and mounted in Fluoromount-G (Southern Biotech, Birmingham, AL). Microscopy was performed on a Nikon Eclipse TE300 inverted microscope equipped with a Nikon 100× objective and a Spot RT Monochrome camera. Digital images were collected by using Spot 4.0 software and saved in TIFF format. Files were opened in Adobe Photoshop CS and cropped to display a single cell. Gray levels were adjusted by using the Auto Levels command with black and white clip both set to 0%. Images then were cropped again and scaled for final display.
Statistics.
All error bars indicate standard deviations and were calculated from triplicate measurements. Where not visible, error bars are smaller than symbols.
Acknowledgments
We thank Donald B. Bloch (Massachusetts General Hospital), Kenneth D. Bloch (Massachusetts General Hospital), and Walther Mothes (Yale University School of Medicine) for kindly sharing cell lines and plasmids. This work was supported by funding from National Institutes of Health Grant R01 DK59934.
Abbreviations
- PMA
phorbol 12-myristate 13-acetate
- FcγRIIA
Fcγ receptor IIa
- RFP
red fluorescent protein.
Footnotes
The authors declare no conflict of interest.
References
- 1.Manz RA, Hauser AE, Hiepe F, Radbruch A. Annu Rev Immunol. 2005;23:367–386. doi: 10.1146/annurev.immunol.23.021704.115723. [DOI] [PubMed] [Google Scholar]
- 2.Bruton OC. Pediatrics. 1952;9:722–728. [Google Scholar]
- 3.Sideras P, Smith CI. Adv Immunol. 1995;59:135–223. doi: 10.1016/s0065-2776(08)60631-8. [DOI] [PubMed] [Google Scholar]
- 4.Vieira OV, Botelho RJ, Grinstein S. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desjardins M, Griffiths G. Curr Opin Cell Biol. 2003;15:498–503. doi: 10.1016/s0955-0674(03)00083-8. [DOI] [PubMed] [Google Scholar]
- 6.Takai T, Li M, Sylvestre D, Clynes R, Ravetch JV. Cell. 1994;76:519–529. doi: 10.1016/0092-8674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 7.Crowley MT, Costello PS, Fitzer-Attas CJ, Turner M, Meng F, Lowell C, Tybulewicz VL, DeFranco AL. J Exp Med. 1997;186:1027–1039. doi: 10.1084/jem.186.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiefer F, Brumell J, Al-Alawi N, Latour S, Cheng A, Veillette A, Grinstein S, Pawson T. Mol Cell Biol. 1998;18:4209–4220. doi: 10.1128/mcb.18.7.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzer-Attas CJ, Lowry M, Crowley MT, Finn AJ, Meng F, DeFranco AL, Lowell CA. J Exp Med. 2000;191:669–682. doi: 10.1084/jem.191.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armstrong JA, Hart PD. J Exp Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones TC, Yeh S, Hirsch JG. J Exp Med. 1972;136:1157–1172. doi: 10.1084/jem.136.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armstrong JA, Hart PD. J Exp Med. 1975;142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joiner KA, Fuhrman SA, Miettinen HM, Kasper LH, Mellman I. Science. 1990;249:641–646. doi: 10.1126/science.2200126. [DOI] [PubMed] [Google Scholar]
- 14.Worth RG, Mayo-Bond L, Kim MK, van de Winkel JG, Todd RF III, Petty HR, Schreiber AD. Blood. 2001;98:3429–3434. doi: 10.1182/blood.v98.12.3429. [DOI] [PubMed] [Google Scholar]
- 15.Stockinger W, Zhang SC, Trivedi V, Jarzylo LA, Shieh EC, Lane WS, Castoreno AB, Nohturfft A. Mol Biol Cell. 2006;17:1697–1710. doi: 10.1091/mbc.E05-12-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stockinger W, Castoreno AB, Wang Y, Pagnon JC, Nohturfft A. J Lipid Res. 2004;45:2151–2158. doi: 10.1194/jlr.D400011-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udenfriend S, Gerber L, Nelson N. Anal Biochem. 1987;161:494–500. doi: 10.1016/0003-2697(87)90479-9. [DOI] [PubMed] [Google Scholar]
- 18.Nimmerjahn F, Ravetch JV. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 19.May RC, Machesky LM. J Cell Sci. 2001;114:1061–1077. doi: 10.1242/jcs.114.6.1061. [DOI] [PubMed] [Google Scholar]
- 20.Underhill DM, Ozinsky A. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 21.Larsen EC, Ueyama T, Brannock PM, Shirai Y, Saito N, Larsson C, Loegering D, Weber PB, Lennartz MR. J Cell Biol. 2002;159:939–944. doi: 10.1083/jcb.200205140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheleznyak A, Brown EJ. J Biol Chem. 1992;267:12042–12048. [PubMed] [Google Scholar]
- 23.Allen LA, Aderem A. J Exp Med. 1996;184:627–637. doi: 10.1084/jem.184.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng Yan Hing JD, Desjardins M, Descoteaux A. Biochem Biophys Res Commun. 2004;319:810–816. doi: 10.1016/j.bbrc.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 25.Kielian MC, Cohn ZA. J Exp Med. 1981;154:101–111. doi: 10.1084/jem.154.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Downey GP, Botelho RJ, Butler JR, Moltyaner Y, Chien P, Schreiber AD, Grinstein S. J Biol Chem. 1999;274:28436–28444. doi: 10.1074/jbc.274.40.28436. [DOI] [PubMed] [Google Scholar]
- 27.Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marme D, Schachtele C. J Biol Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 28.Jahraus A, Egeberg M, Hinner B, Habermann A, Sackman E, Pralle A, Faulstich H, Rybin V, Defacque H, Griffiths G. Mol Biol Cell. 2001;12:155–170. doi: 10.1091/mbc.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjeken R, Egeberg M, Habermann A, Kuehnel M, Peyron P, Floetenmeyer M, Walther P, Jahraus A, Defacque H, Kuznetsov SA, Griffiths G. Mol Biol Cell. 2004;15:345–358. doi: 10.1091/mbc.E03-05-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbas AK, Lichtman AH. Cellular and Molecular Immunology. Philadelphia: WB Saunders; 2003. [Google Scholar]
- 31.Aslam M, Dent AH. Bioconjugation: Protein Coupling Techniques for the Biomedical Sciences. London: Macmillan; 1998. [Google Scholar]
- 32.Artavanis-Tsakonas K, Love JC, Ploegh HL, Vyas JM. Proc Natl Acad Sci USA. 2006;103:15945–15950. doi: 10.1073/pnas.0607528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loos H, Blok-Schut B, van Doorn R, Hoksbergen R, Brutel de la Riviere A, Meerhof L. Blood. 1976;48:731–742. [PubMed] [Google Scholar]
- 34.Graham FL, Smiley J, Russell WC, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 35.Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann MC, Narisawa S, Hess RA, Millan JL. Exp Cell Res. 1992;201:417–435. doi: 10.1016/0014-4827(92)90291-f. [DOI] [PubMed] [Google Scholar]
- 37.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- 38.Collins SJ, Gallo RC, Gallagher RE. Nature. 1977;270:347–349. doi: 10.1038/270347a0. [DOI] [PubMed] [Google Scholar]
- 39.Raschke WC, Baird S, Ralph P, Nakoinz I. Cell. 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 40.Ralph P, Nakoinz I. Nature. 1975;257:393–394. doi: 10.1038/257393a0. [DOI] [PubMed] [Google Scholar]
- 41.Sundstrom C, Nilsson K. Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]