Abstract
Excessive activation of the nuclear enzyme, poly(ADP-ribose) polymerase-1 (PARP-1) plays a prominent role in various of models of cellular injury. Here, we identify poly(ADP-ribose) (PAR) polymer, a product of PARP-1 activity, as a previously uncharacterized cell death signal. PAR polymer is directly toxic to neurons, and degradation of PAR polymer by poly(ADP-ribose) glycohydrolase (PARG) or phosphodiesterase 1 prevents PAR polymer-induced cell death. PARP-1-dependent, NMDA excitotoxicity of cortical neurons is reduced by neutralizing antibodies to PAR and by overexpression of PARG. Neuronal cultures with reduced levels of PARG are more sensitive to NMDA excitotoxicity than WT cultures. Transgenic mice overexpressing PARG have significantly reduced infarct volumes after focal ischemia. Conversely, mice with reduced levels of PARG have significantly increased infarct volumes after focal ischemia compared with WT littermate controls. These results reveal PAR polymer as a signaling molecule that induces cell death and suggests that interference with PAR polymer signaling may offer innovative therapeutic approaches for the treatment of cellular injury.
Keywords: excitotoxicity, poly(ADP-ribose) glycohydrolase, poly(ADP-ribose) polymerase, stroke
Poly(ADP-ribose) polymerase-1 (PARP-1) is an abundant nuclear protein that is involved in the DNA base excision repair system, where it is potently activated by DNA strand nicks and breaks (1, 2). Using NAD+ as a substrate, PARP-1 builds up homopolymers of ADP ribose units on various nuclear proteins including histones, DNA polymerases, topoisomerases, DNA ligase-2, transcription factors (3, 4), and PARP-1 itself (5, 6). Although the exact physiologic function of PARP-1 is not completely understood, in some tissues it plays an important role in DNA repair and genomic stability (5, 7, 8). Poly(ADP-ribose) (PAR) catabolism and metabolism is a dynamic process, with PAR glycohydrolase (PARG) playing the major role in the degradation of the polymer (9).
Recent studies using pharmacologic inhibition of PARP or genetic KO of PARP-1 indicate that PARP-1 plays a dramatic and significant role in cellular injury after stroke, trauma, ischemia–reperfusion of the heart, spleen, skeletal muscle, and retina, arthritis, β-islet cytotoxicity/diabetes mellitus, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) model of Parkinson's disease, experimental autoimmune encephalomyelitis (EAE) model of multiple sclerosis, endotoxic shock, multiple-system organ failure, and liver damage (for review, see refs. 1 and 10). PARP-1 activation also plays a prominent role in NMDA excitotoxicity, because PARP-1 KO mice are remarkably resistant both in vitro and in vivo to the excitotoxic effects of glutamate and NMDA (11, 12). A cell-suicide hypothesis has been proposed (1, 2, 13, 14) to explain the actions of PARP-1 in mediating cell death. However, studies in mice lacking PARG suggest that PAR polymer formed during the activation of PARP-1 might play a role in PARP-1-dependent cell death. PARG KO mice die at embryonic day 3.5 because of the failure to hydrolyze PAR polymer and its subsequent accumulation (15). Moreover, the loss of PARG in Drosophila melanogaster leads to lethality in the larval stage at a normal developmental temperature (25°C) and progressive neurodegeneration with reduced locomotor activity and a short lifespan at 29°C because of the accumulation of PAR polymer (16). Here, we explore the role of PAR polymer in PARP-1-dependent cell death and report that PAR polymer is a cell-death signal that plays an important role in PARP-1-dependent cell death.
Results
PAR Polymer Is Toxic.
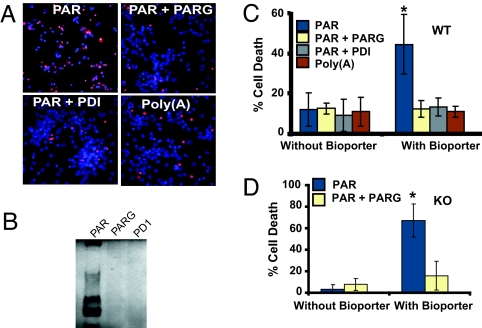
An extensive amount of PAR polymer is formed, and it remains significantly elevated for a prolonged period after a variety of toxic insults in which PARP-1 activation plays a prominent role, (12, 17–19). To determine whether PAR polymer is sufficient to induce neuronal cell death, viability was monitored by Hoechst 33342 and propidium iodide (PI) staining by using quantitative computer-assisted counting of viable and dead neurons (20) after BioPorter-mediated delivery of PAR polymer. BioPorter reagent is a cationic lipid formulation TFA-DODAP:DOPE that enables delivery of recombinant proteins, peptides, or antibodies into viable cells (21). The PAR polymer was synthesized by in vitro automodification of PARP-1 and has a mean length of 40 ADP-ribose residues, as determined by HPLC methods and gel electrophoresis (22), and, accordingly, the concentration of PAR is given as a function of polymer molecules with a mean size of 40 ADP-ribose units. The range of size of PAR in this mix is 6- through 100-mer ADP-ribose units (22, 23). PAR polymer is effectively delivered into cortical neurons by the BioPorter reagent, as revealed by using PAR polymer antibody and confocal imaging. PAR polymer is observed in the majority of neurons (Fig. 5, which is published as supporting information on the PNAS web site). Under these conditions, PAR polymer induces ≈50% cell death in cortical neurons, which is prevented by pretreatment of the PAR polymer with PARG or phosphodiesterase 1 (PD1), which degrade PAR polymers [Fig. 1B (24, 25)] (Fig. 1 A and C). Because PAR polymer is a highly negatively charged molecule, the same concentration of poly (adenine) [poly(A)], also negatively charged, was applied by BioPorter to cortical neurons. BioPorter-mediated delivery of poly(A) fails to elicit neuronal cell death (Fig. 1 A and C). To determine whether PAR polymer induces cell death in the absence of PARP-1, PAR polymer was delivered to PARP-1 KO cortical neurons, and cell death was monitored (Fig. 1D). PAR polymer delivery induces cell death in PARP-1 KO cortical neurons that is attenuated by pretreatment with PARG (Fig. 1D). These results taken together indicate that PAR polymer is toxic and that PAR polymer can act as a death signal in PARP-1 KO cultures.
Fig. 1.
PAR polymer induces neuronal cell death. (A) Representative fluorescent microscopic images of PI (red) and Hoechst (blue) staining after treatment of neurons with BioPorter-mediated delivery of PAR polymer, PAR polymer + PARG, PAR polymer + PD1, and poly(A). (B) Silver staining of purified PAR polymer, showing degradation of PAR polymer after incubation with PARG (PAR+PARG) and PD1 (PAR+PD1). These experiments (A and B) have been replicated in separate experiments at least three times with similar results. (C) Quantitative analysis of Hoechst- and PI-stained WT neurons after BioPorter-mediated delivery of PAR polymer, PAR polymer + PARG, PAR polymer + PD1, and poly(A). Purified PAR, cleaved PAR by either PARG or PD1 pretreatment, or synthesized poly(A) was diluted with PBS, mixed with BioPorter reagent, and added to neuronal cultures at a final concentration of 80 nM in serum-free MEM. The neurons were cultured for 3 h to allow the polymers to be imported into neurons and then subjected to fluorescent microscopy and quantitative computer-assisted cell counting after Hoechst and PI double staining 24 h later. Data are the mean ± SD, n = 3; ∗, P < 0.05. (D) PAR polymer can also induce cell death in PARP-1 KO neurons. PAR polymer (80 nM) and cleaved PAR (80 nM PAR polymer + PARG) was delivered into neurons by BioPorter-mediated delivery, and cell death was analyzed by Hoechst and PI staining. Data are the mean ± SD, n = 3; ∗, P < 0.05.
High-Molecular-Weight PAR Polymers Are Toxic.
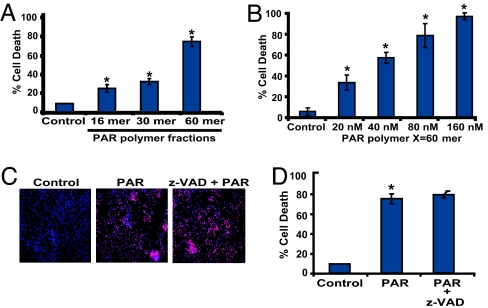
These latter studies were performed with a mixture of PAR polymers of different size and complexity. To determine the nature of the PAR polymer that mediates cell death, the ability of isolated PAR polymers to induce cell death was evaluated by using PAR polymers of different size and complexity, ranging in length from 16 to >60 ADP-ribose residues. The DEAENPR fractions, as defined by Kiehlbauch and Jacobson (23), were used for these studies. Increasing length and complexity of PAR polymers leads to increased cell death, with complex polymers >60 ADP-ribose units inducing >80% cell death at a concentration of 80 nM polymer (Fig. 2A). At an equivalent polymer concentration, polymers of 16 ADP-ribose residues elicit a small amount of cell death, and polymers of 30 ADP-ribose residues induce modest cell death (Fig. 2A). Thus, consistent with formation of high-molecular-weight complex polymers after PARP-1-dependent cell death (12, 18, 19), PAR polymers of increasing complexity and molecular weight are more toxic. A dose–response relationship of complex polymers >60 ADP-ribose units on cortical neuron cell death was performed. Concentrations as low as 20 nM PAR polymer induce cell death, and cell death increases with increasing concentration of PAR polymer (Fig. 2B). These concentrations, length, and complexity of PAR polymers are within the range of polymer concentrations and size found in intact cells during NMDA excitotoxicity (12, 18, 19) (see Fig. 4), and PARP-1-dependent [N-methyl-N-nitro-N-nitrosoguanidine (MNNG)] HeLa cell toxicity (data not shown). Thus, it is likely that complex and high-molecular-weight PAR polymers act, in part, to play a role of PARP-1 in cell death.
Fig. 2.
PAR polymer toxicity is length- and dose-dependent and caspase-independent. (A) Purified PAR polymer was fractionated into fractions of varying polymer length. Different PAR polymer fractions with average polymer size (16, 30, and 60 mer) were delivered into the cortical neurons by the BioPorter delivery reagent at a final concentration of 80 nM. The most abundant fraction containing PAR polymer of 40–100 (average polymer size 60 mer) ADP-ribose residues induces robust neuronal cell death. Neurons were treated with BioPorter alone and serve as a control. Data are the mean ± SEM, n = 6; ∗, P < 0.05. (B) PAR polymer-induced neuronal death is dose-dependent. Different concentrations of PAR polymer (20–160 nM) were administered to cortical neurons by the BioPorter reagent, and cell death was assessed by Hoechst and PI staining. Data are the mean ± SD, n = 4; ∗, P < 0.05. (C) Representative pictures of PI- (red) and Hoechst- (blue) stained neurons were treated with BioPorter alone (Control), purified PAR polymer (PAR), or purified PAR polymer in the presence of z-VAD (z-VAD + PAR), by using BioPorter reagent as a delivery system. PI-positive cells were considered dead cells. (D) Quantification of the PAR polymer-mediated cell death in the presence of z-VAD. z-VAD is unable to protect against PAR polymer-mediated cell death in cortical neurons. BioPorter alone serves as a control. Cell death was assessed by Hoechst and PI staining. Data are the mean ± SEM, n = 6; ∗, P < 0.05.
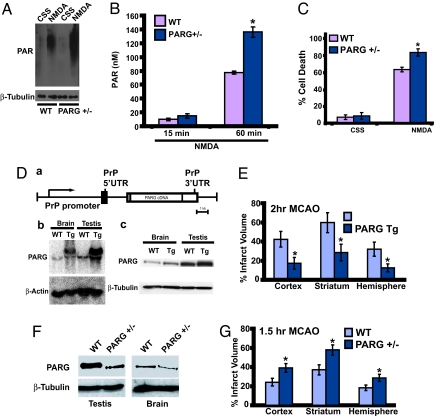
Fig. 4.
Enhanced PAR polymer formation and toxicity in PARG+/− mice, and PAR polymer mediates PARP-1-dependent cell death in vivo. (A) Western blots showing PAR polymer formation in mouse cortical neurons prepared from PARG+/− mice and their WT littermates. The neurons were subjected to NMDA stimulation for 5 min on DIV 14. Samples were harvested 1 h after NMDA stimulation and probed for PAR polymer by using anti-PAR antibody. PARG+/− neurons that express reduced levels of PARG accumulate more PAR polymer than their WT littermates. PARG+/− neurons also have higher basal PAR polymer levels. The same blot was stripped and probed for β-tubulin, serving as a loading control. This experiment has been replicated in separate experiments at least three times with similar results. (B) Cortical neurons from PARG+/− KO mice and WT littermate cultures were treated with 500 μM NMDA for 5 min on DIV 14, and intracellular PAR polymer levels were determined at the indicated times points after NMDA administration by using an ELISA as described in Materials and Methods. Data are the mean ± SD, n = 4; ∗, P < 0.05. (C) Cortical neurons from PARG+/− KO mice and WT littermate cultures were subjected to NMDA-stimulation (500 μM for 5 min) on DIV 14. PARG+/− neurons that have reduced expression of PARG are more sensitive to NMDA toxicity than their WT littermates. Control cultures were treated with control salt solution (CSS) alone. Data are the mean ± SD, n = 6; ∗, P < 0.05. (D) PARG Tg mice overexpress PARG. (a) Expression of the PARG gene is under the control of the PrP promoter. (b) Northern blot analysis for PARG in WT vs. PARG Tg mice. β-actin was used as a loading control. (c) Immunoblotting of mouse PARG in whole-brain and testis homogenates of transgenic mice expressing the PARG and WT control mice. β-tubulin was used as a loading control. (E) Infarct volume (±SEM) in cerebral cortex, striatum, and total hemisphere (expressed as a percent of contralateral structure after correction for swelling) in 10 WT and 11 PARG Tg mice subjected to 120 min of MCAO. ∗, P < 0.05 between groups. (F) Decreased levels of PARG protein in PARG+/− mice. Immunoblot analysis of full-length (110-kDa) PARG protein levels in brain and testis extracts from PARG+/+ and PARG+/− mice. These results have been replicated at least two separate times with similar results. (G) Infarct volume (± SEM) in cerebral cortex, striatum, and total hemisphere (expressed as a percent of contralateral structure after correction for swelling) in 15 WT and 16 PARG+/− mice subjected to 90 min of MCAO. ∗, P < 0.05 between groups.
PAR Polymer-Induced Cell Death Is Caspase-Independent.
PARP-1-dependent cell death is, in part, caspase-independent (18, 19). To determine whether PAR polymer-induced cell death is caspase-independent, we monitored cell death of cortical neurons after BioPorter-mediated delivery of PAR polymer in the presence and absence of the broad-spectrum caspase inhibitor Z-VAD.fmk. Z-VAD fails to prevent PAR polymer-induced cell death (Fig. 2 C and D). PAR polymer also fails to directly activate caspases (data not shown). To ascertain whether these findings extend to nonneuronal cells and whether PAR polymer death signaling occurs outside the nervous system, we evaluated PAR toxicity in HeLa cells. Similar to neurons, complex polymers, >60 ADP-ribose units, cause HeLa cell death, with 70% cell death at a PAR polymer concentration of 80 nM (Fig. 6A, which is published as supporting information on the PNAS web site). Z-VAD also fails to prevent PAR polymer-induced cell death in HeLa cells (Fig. 6B). Taken together, these results indicate that PAR polymer induces cell death in a concentration-dependent and caspase-independent manner.
PAR Polymer Mediates PARP-1-Dependent Cell Death.
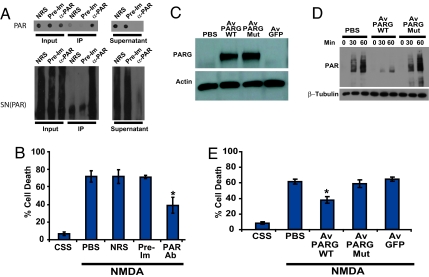
To determine what role PAR polymer formation might play in NMDA excitotoxicity, a pathophysiologically relevant form of PARP-1-dependent cell death, a neutralizing polyclonal antiserum to PAR polymer was used (Fig. 3). Anti-PAR antiserum is capable of immunodepleting both purified PAR polymer and PARP-1-activated nuclear supernatant containing endogenous PAR polymer [SN(PAR)], as assessed by dot–blot analysis and Western blot analysis, respectively (Fig. 3A). Anti-PAR antiserum was delivered into cortical neurons by the BioPorter reagent before NMDA treatment. With this protein-delivery system, >95% of the neurons receive anti-PAR antiserum, as indicated by the presence of a fluorescent marker (data not shown), which was coadministered with the antiserum. BioPorter-mediated delivery of anti-PAR antiserum reduces NMDA excitotoxicity, whereas BioPorter-mediated delivery of an equivalent concentration of preimmune serum or normal rabbit serum fails to protect neurons from NMDA-induced cell death (Fig. 3B). Similar neuroprotection against NMDA toxicity was obtained with a different anti-PAR antiserum (data not shown). To further evaluate the role of PAR polymer in NMDA-induced excitotoxicity, a PARG adenovirus was constructed by using the exon-1-deleted form of PARG, which is exclusively localized to the cytoplasm (Av PARG WT) (26). In addition, we constructed a catalytically inactive exon-1-deleted mutant PARG adenovirus (Av PARG Mut) in which two critical glutamates at residues 756 and 757 were mutated to alanines (27). Adenoviral-mediated overexpression of WT and mutant PARG was assessed by Western blot analysis, and we observe a several-fold increase in the level of both WT and mutant exon-1-deleted PARG (Fig. 3C). Adenoviral-mediated overexpression of WT cytosolic PARG leads to a reduction of cytosolic PAR polymer (Fig. 3D) and an ≈50% reduction of NMDA-induced neuronal cell death (Fig. 3E), whereas overexpression of the catalytically inactive mutant PARG fails to reduce cytosolic PAR polymer levels (Fig. 3D) and NMDA toxicity (Fig. 3E). To control for nonspecific effects of adenoviral-mediated gene expression, adenoviral-mediated overexpression of GFP was evaluated and was found to have no effect on NMDA excitotoxicity (Fig. 3E).
Fig. 3.
Interfering with PAR signaling reduces NMDA-induced neuronal cell death. (A) Immunodepletion of purified PAR polymer (Upper) and SN(PAR) (Lower) by anti-PAR antibody. Supernatants after immunoprecipitation have reduced level of PAR polymers. NRS, normal rabbit serum; Pre-Im, preimmune serum; α-Par, anti-Par antibody. (B) Pretreatment with neutralizing PAR antiserum reduces NMDA-induced neuronal cell death. PAR antiserum, normal rabbit serum, or preimmune serum was delivered into cortical neurons with the BioPorter reagent. At 3 h after delivery, the neurons were treated with NMDA (500 μM for 5 min). Cell death was assessed 18–24 h later by Hoechst and PI staining. Data are the mean ± SEM, n = 6–8; ∗, P < 0.05. (C) Cortical neurons were infected with a cytosolic form (exon-1 deleted) PARG adenoviral construct (Av PARG WT) or catalytically inactive (Av PARG Mut) virus, and, after 48 h, the cells were harvested in lysis buffer containing protease inhibitors. Because of low basal levels of PARG, nondetectable levels of PARG are seen in adeno-GFP- and PBS-treated cultures, whereas high levels of PARG expression are seen in cultures that are treated with Av PARG WT or Av PARG Mut virus. (D) Overexpression of cytosolic, WT PARG decreases PAR levels in cortical neurons after NMDA treatment. Neurons [11 days in vitro (DIV)] were infected with an exon 1-deleted, cytosolic WT (Av PARG WT) and mutant (Av PARG mut) PARG adenoviral constructs, or a GFP-adenoviral control construct (Av GFP). After 48 h, the transduced cultures were treated with NMDA (500 μM for 5 min), and neurons were harvested at indicated time points and subjected to Western blotting analysis with anti-PAR antibody. These experiments (A, C, and D) have been replicated in separate experiments at least three times with similar results. (E) Overexpression of PARG reduces NMDA-induced neuronal cell death. Neurons (12 DIV) were infected with a Av PARG WT or catalytically inactive Av PARG Mut virus, GFP-adenoviral control construct (Adeno-GFP), or no-adenovirus (PBS). After 48 h, the neurons were treated with NMDA (500 μM for 5 min), and 18–24 h after NMDA administration, cells were stained with Hoechst and PI to assess cell death. Data are the mean ± SEM, n = 6; ∗, P < 0.05.
To determine whether the anti-PAR antibody and adenoviral-mediated overexpression of WT cytosolic PARG could block against other forms of PARP-1-dependent cell death in nonneuronal cells, we assessed the ability of the neutralizing PAR antiserum and adenoviral-mediated overexpression of WT cytosolic PARG to block PARP-1-dependent cell death induced by MNNG in HeLa cells. HeLa cells die in a PARP-1-dependent/caspase independent manner after MNNG treatment, because treatment of HeLa cells with 50 μM MNNG for 15 min leads to cell death that is blocked by the PARP inhibitor, 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone (DPQ) (30 μM), but the pancaspase inhibitor z-VAD (100 μM) has no effect (Fig. 7A, which is published as supporting information on the PNAS web site). BioPorter-mediated delivery of anti-PAR antibody reduces MNNG cell death, whereas a preimmune serum and normal rabbit serum have no effect and adenoviral-mediated overexpression of WT cytosolic PARG reduces MNNG induced cell death, whereas control virus has no effect (Fig. 7B). The residual toxicity observed with reducing PAR polymer levels in neurons and HeLa cells may be due to alternative pathways of cell death or residual PAR polymer, because our methods of depleting PAR polymer are not complete. Consistent with this notion is our observation that concentrations of PAR polymer as low as 20 nM are still toxic (see Fig. 2B).
To ascertain whether reducing the level of PARG leads to enhanced PARP-1-dependent cell death, we evaluated NMDA excitotoxicity in mice lacking one copy of the PARG gene. PARG KO mice die at embryonic day 3.5, but the heterozygotes are viable (15). We first determined whether there is increased PAR polymer formation in PARG+/− cortical neurons compared with WT cortical neurons after a toxic dose of NMDA (Fig. 4A and B). PARG+/− cortical neurons have enhanced PAR polymer formation after NMDA administration, as assessed by Western blot analysis with an anti-PAR antibody (Fig. 4A). Quantitation of the amount of PAR polymer formed in WT cultures indicates that ≈80 nM of PAR polymer is detected at 60 min after NMDA administration (Fig. 4B). There is almost a 2-fold increase in PAR polymer in PARG+/− mice cortical neurons compared with WT cortical neurons (Fig. 4B). We next examined neuronal cell death and found that there is an ≈20% increase in cell death after NMDA administration in PARG+/− cortical neurons compared with WT cortical neurons (Fig. 4C). The 20% increase in cell death in PARG+/− cortical neurons after NMDA toxicity is essentially identical to the percentage increase in cell death after a comparable increase of the concentration of BioPorter-mediated delivery of purified PAR polymer (see Fig. 2B).
To determine whether reducing PARG levels in HeLa cells increases their susceptibility to PARP-1-dependent cell death, HeLa cells were treated with an siRNA to PARG (Fig. 8A, which is published as supporting information on the PNAS web site). siRNA treatment of HeLa cells leads to a significant reduction in PARG catalytic activity, as assessed by a PARG activity zymogram activity assay (Fig. 8A), and protein, as assessed by Western blot analysis (Fig. 8A). PARG siRNA leads to enhanced PAR polymer formation after MNNG treatment (Fig. 8B). In mock and scrambled siRNA-transfected cells, 50 μM MNNG for 15 min induces transient PAR polymer formation (Fig. 8B). PARG siRNA treatment enhances the amount of PAR polymer detected after MNNG treatment of HeLa cells (Fig. 8B). PAR polymer accumulation is substantially enhanced after PARG siRNA treatment but eventually is degraded by the residual PARG activity (Fig. 8B). Under conditions in which PARG siRNA reduces PARG activity, we assessed MNNG toxicity in HeLa cells (Fig. 8C). MNNG (50 μM) for 15 min leads to killing of ≈50% of the HeLa cells, and PARG siRNA treatment enhances MNNG-induced cell death by 30% (Fig. 8B). Taken together, these data generated from different and diverse experiments are consistent with PAR polymer acting as a signal that initiates the cell-death cascade after PARP-1-dependent cell death.
PAR Polymer Mediates Neuronal Injury in Vivo.
To evaluate whether PAR polymer formation plays a role in the susceptibility of neurons to a pathophysiologically important insult in vivo, PARG transgenic mice overexpressing WT PARG under the direction of the mouse prion promoter in which PARG message and protein are overexpressed (Fig. 4D) and PARG+/− mice, which have reduced levels of PARG protein (Fig. 4F), were subjected to transient middle cerebral artery occlusion (MCAO) by the intralumenal filament technique as described by Goto et al. (28). The effect of overexpression of PARG on focal ischemic injury was tested by comparing infarct volume after 120 min of transient MCAO in PARG transgenic (Tg) mice and WT littermates. For the PARG transgenic studies we used aged-matched littermate controls from crosses between PARG heterozygote Tg and WT mice; thus, all mice used in these studies should have similar genetic backgrounds. Induction of MCAO reduced cortical perfusion in the MCA territory measured by laser-Doppler flowmetry (LDF) to an equivalent extent in WT (15 ± 7% of baseline; n = 10) and PARG Tg (18 ± 5%; n = 11) mice. Rectal temperature was similar between groups at 10 min of MCAO (WT 36.9 ± 0.5°C vs. PARG Tg 37.0 ± 0.6°C) and at the end of MCAO (WT 36.2 ± 0.1°C vs. PARG Tg 36.2 ± 0.2°C). The neurologic-deficit score was similar in WT (2.4 ± 0.3) and PARG Tg (2.8 ± 0.3) during MCAO. After transient MCAO, overall infarct volume in the hemisphere of PARG Tg mice is significantly reduced, by 62%, compared with WT littermates (Fig. 4E). On a regional basis, infarct volume is reduced by 52% in striatum and by 59% in cerebral cortex of PARG Tg mice. The effect of decreasing PARG expression (Fig. 4F) on focal ischemic injury was next tested on PARG+/− mice. For the PARG+/− studies, we used aged-matched littermate controls from crosses between PARG+/− mice; thus, all mice used in these studies should have similar genetic backgrounds. The reduction in LDF during MCAO in PARG+/− mice (18 ± 7% of baseline; n = 15) was not different from that in WT littermates (16 ± 6%; n = 16). Rectal temperature was similar between groups at 10 min of MCAO (WT 37.0 ± 0.6°C vs. PARG+/− 37.3 ± 0.3°C) and at the end of MCAO (WT 36.2 ± 0.2°C vs. PARG Tg 36.2 ± 0.2°C). During MCAO, the neurologic-deficit scores were equivalent (WT 2.9 ± 0.6 vs. PARG+/− 2.7 ± 0.4). After 90 min of transient MCAO, infarct volume is significantly increased, by 62%, in cerebral cortex, 56% in striatum, and 56% in the entire hemisphere of PARG+/− mice compared with WT littermates (Fig. 4G). These results taken together suggest that PAR polymer is the nuclear signal initiating the cell-death cascade after PARP-1-dependent cell death in vivo.
Discussion
The major finding of this article is that PAR polymer acts as a previously uncharacterized signaling molecule that induces cell death after PARP-1 activation. Evidence supporting PAR polymer as a cell-death signaling molecule include the observations that PAR polymer is directly toxic and that its toxicity is abolished by pretreatment of the PAR polymer with either PD1 or PARG, which degrade PAR polymer. Moreover, interfering with PAR polymer signaling through neutralizing PAR antibodies or PARG overexpression reduces PARP-1-dependent NMDA excitotoxicity and MNNG-induced cell death. In addition, Tg mice overexpressing PARG have markedly reduced infarct volumes after 2 h of transient MCAO. Consistent with the notion that PAR polymer is toxic are our observations that PARG heterozygote KO cortical cultures are more sensitive to NMDA excitotoxicity, siRNA knockdown of PARG leads to enhanced MNNG toxicity, and PARG heterozygote KO mice have larger infarct volumes after 1.5 h of transient MCAO.
During PARP-1-dependent cell death, an extensive amount of PAR polymer is formed through consumption of intracellular NAD+ stores. Energy depletion was thought to be the primary mechanism by which excessive PARP-1 activation kills cells (10). Although we cannot exclude the possibility that decrements in NAD+ contribute to cell death, because sustained energy depletion clearly can kill cells, the ability of neutralizing PAR antibodies and overexpression of PARG to reduce NMDA excitotoxicity and MNNG toxicity, the reduction in focal ischemia-induced neuronal injury by PARG overexpression and the direct toxicity of PAR polymer in energy-replete cells supports the idea that NAD+ decrements are not sufficient for PARP-1-dependent cell death. Consistent with this notion are studies showing that energy depletion alone may be an insufficient explanation for why PARP-1 activation kills cells in vivo (28).
In summary, our results suggest that excessive activation of PARP-1 leads to a unique intrinsic cell-death program and that PAR polymer participates in PARP-1-dependent cell death. Interfering with PAR actions may offer innovative therapeutic approaches to treat cellular injury.
Methods
Primary Cortical Cultures.
Primary cortical neuron cultures were prepared from gestational day 14–15 fetal mice as described (20), and, in mature cultures, neurons represent 80–90% of the total number of cells (29).
PAR Polymer Synthesis and Purification.
Purification and synthesis of PAR polymer were performed as described (23, 30)
BioPorter Protein-Delivery System.
BioPorter reagent was purchased from Gene Therapy Systems (San Diego, CA) (21). Antibodies, PAR polymer, or cleaved PAR polymer with either PD1 (Sigma, St. Louis, MO) or recombinant PARG was diluted to desired concentration with PBS. The diluted protein solution was added to the dried BioPorter reagent and allowed to rest at room temperature for 5 min, followed by gentle mixing. Serum-free medium was added to bring the BioPorter/PAR or BioPorter protein complex up to 250–300 μl. After a wash in serum-free media, the cell cultures were incubated with the BioPorter/PAR or BioPorter/protein complex for 3–4 h at 37°C. Cultures were subsequently used for experiments.
Cytotoxicity.
Neurons (14 DIV) were exposed to PAR polymer or to NMDA (20). Viability was determined by computer-assisted cell counting after staining of all nuclei with 7 μM Hoechst 33342 (Invitrogen, Carlsbad, CA) and dead cell nuclei with 2 μM PI (Invitrogen). The numbers of total and dead cells were counted with the Axiovision 4.3 software (Zeiss, Thornwood, NY).
Western Blotting.
Cell lysates or subcellular fractions were size-separated through SDS/PAGE and processed for analysis by using a Supersignal ChemiLuminiscence detection kit (Pierce, Rockford, IL) as described by the manufacturer (18). All primary antibodies are previously characterized, including anti-PAR polyclonal antibody (30) and rabbit anti-poly(ADP-ribose) glycohydrolase polyclonal antibody (15).
Transduction of Cultured Neurons with Recombinant Adenovirus.
An exon-1-deleted WT PARG gene was subcloned into an adenoviral vector, and its expression was driven by a CMV promoter. To make a catalytically inactive mutant construct, two adjacent glutamic residues at 756 and 757 were point-mutated to alanines, and the exon-1-deleted mutant PARG gene was cloned in the same adenoviral vector as the exon-1-deleted WT gene. A GFP adenovirus was used as a control. The viral constructs were diluted with MEM containing 2% serum and 21 mM glucose and added onto cultured neurons (12 DIV) or HeLa Cells. After 24 h, the medium was changed into 10% serum-containing medium, and then the cells were cultured for an additional day to allow PARG or GFP expression. After 2 days, the cells were harvested for Western blot analysis or subjected to NMDA administration. Cell death was detected after the insult by Hoechst 33342 and PI double staining and computer-assisted quantification.
Isolation and ELISA of Intracellular PAR Polymer.
Preparation and quantification of intracellular PAR polymer from cortical neurons were performed as described (31). PAR polymer standards used in ELISA were a complex form (average 60-mer ADP-ribose). Intracellular PAR polymer concentration was calculated based on the assumption that the diameter of neurons is 16.78 μm (32).
Focal Cerebral Ischemia.
All procedures on mice were preapproved by the Johns Hopkins University Animal Care and Use Committee and conformed with the principles of laboratory animal research of the American Physiological Society. Transient MCAO was produced in mice by the intralumenal-filament technique as described (28). Mice were anesthetized with 5% halothane and maintained on 1–1.5% halothane in O2-enriched air by using a face mask. Rectal temperature was maintained at 36–37°C with a warming blanket and heating lamp. MCAO was produced by inserting a 6–0 nylon monofilament, with the tip coated by cyanoacrylate glue, into the internal carotid artery through a cut stump of the external carotid artery while the common carotid artery was occluded. A laser Doppler flowmetry probe placed on the skull over the lateral parietal cortex was used to verify MCAO over the first 10 min of occlusion. Anesthesia was discontinued, and a neurological examination was performed to verify functional deficits on a scale of 0–4: 0 = no deficit; 1 = forelimb weakness; 2 = circling to affected side; 3 = unable to bear weight on affected side; and 4 = no spontaneous motor activity (33). To allow reperfusion, the mouse was briefly anesthetized with halothane, and the filament was withdrawn. The brain was harvested 1 day after MCAO for analysis of infarct volume by triphenyltetrazolium chloride staining of five coronal sections. Infarct size was expressed as a percent of the contralateral structure after correcting for swelling (34).
Mouse Strains.
PARP-1 KO and PARG KO mice have been previously described. PARP-1 KOs are maintained on an outbred strain of 129 SvEv, and 129 SvEv mice were used as controls (12). PARG KO mice are embryonic lethals and, for these studies, PARG+/− mice were crossed, and PARG+/− and age-matched littermate controls were used (15). PARG transgenic mice were generated by creating a transgenic expression construct containing the cDNA of mouse PARG (gift from M. K. Jacobson, University of Arizona, Tucson, AZ) driven by the mouse PrP promoter (PrP-musPARG). A XhoI-SalI fragment containing mouse PARGcDNA was subcloned into the XhoI cloning site of MoPrP to yield PrP-musPARG. The purified DNA fragment of PrP-musPARGwas microinjected into pronuclei of oocyte from a F1 hybrid of C57BL/6J × SJL/J. The presence or absence of the transgene in the resulting animals and their progeny was determined by using tail DNA. The primers PrP-S = 5′-CCTCTTTGTGACTATGTGGA-CTGATGTCGG-3′, PrP-AS = 5′-GTGGATACCCCCTCCCC-CAGCCTAGACC-3′, and PARG3′ = 5′-CCAGCGCTATTTT-AGAAGCTGGCCAAGAGG-3′ were used for PCR genotyping. The following PCR protocol was used: 94°C for 4.5 min, 30 cycles of 94°C for 1.5 min, 72°C for 2 min, and a final extension at 72°C for 4 min.
Northern Blot Analysis.
Total RNA was isolated with TRIzol (Invitrogen) following the manufacturer's instructions. Twenty micrograms of total RNA was resolved in 1% agarose gel by using a NorthernMax-Gly kit (Ambion, Austin, TX) and transferred onto the nylon membrane (Hybond-N; Amersham, Piscataway, NJ) and exposed to a phosphor screen (Perkin Elmer, Meriden, CT) overnight, and the screen was scanned with the cyclone imaging system as described (15).
Immunohistochemistry and HeLa Cell Experiments.
See Supporting Methods, which is published as supporting information on the PNAS web site.
Statistical Analysis.
Statistical ANOVA was applied, followed by the Turkey multiple-comparison test. Data are shown as mean ± SD or SEM; P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Ashok B. Ramalingam and Marc F. Poitras for the initial characterization of PARG transgenic mice, Suk Jin Hong for assistance with adeno-PARG viruses, and Sika Zheng for assistance with adeno-GFP virus. This work was supported by National Institutes of Health Grant NS39148 and the American Heart Association. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Abbreviations
- DIV
days in vitro
- MCAO
middle cerebral artery occlusion
- MNNG
N-methyl-N-nitro-N-nitrosoguanidine
- PAR
poly(ADP-ribose)
- PARG
PAR glycohydrolase
- PARP-1
PAR polymerase-1
- PD1
phosphodiesterase 1
- poly(A)
poly (adenine)
- Tg
transgenic.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Yu SW, Wang H, Dawson TM, Dawson VL. Neurobiol Dis. 2003;14:303–317. doi: 10.1016/j.nbd.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Szabo C, Dawson VL. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 3.Shall S, de Murcia G. Mutat Res. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 4.Smulson ME, Simbulan-Rosenthal CM, Boulares AH, Yakovlev A, Stoica B, Iyer S, Luo R, Haddad B, Wang ZQ, Pang T, et al. Adv Enzyme Regul. 2000;40:183–215. doi: 10.1016/s0065-2571(99)00024-2. [DOI] [PubMed] [Google Scholar]
- 5.Oliver FJ, Menissier-de Murcia J, de Murcia G. Am J Hum Genet. 1999;64:1282–1288. doi: 10.1086/302389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Murcia G, Schreiber V, Molinete M, Saulier B, Poch O, Masson M, Niedergang C, Menissier de Murcia J. Mol Cell Biochem. 1994;138:15–24. doi: 10.1007/BF00928438. [DOI] [PubMed] [Google Scholar]
- 7.Nicoletti VG, Stella AM. Neurochem Res. 2003;28:187–194. doi: 10.1023/a:1022316914492. [DOI] [PubMed] [Google Scholar]
- 8.Hassa PO, Hottiger MO. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. J Biol Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 10.Virag L, Szabo C. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 11.Eliasson MJ, Sampei K, Mandir AS, Hurn PD, Traystman RJ, Bao J, Pieper A, Wang ZQ, Dawson TM, Snyder SH, Dawson VL. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 12.Mandir AS, Poitras MF, Berliner AR, Herring WJ, Guastella DB, Feldman A, Poirier GG, Wang ZQ, Dawson TM, Dawson VL. J Neurosci. 2000;20:8005–8011. doi: 10.1523/JNEUROSCI.20-21-08005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger NA. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 14.Yamamoto H, Uchigata Y, Okamoto H. Nature. 1981;294:284–286. doi: 10.1038/294284a0. [DOI] [PubMed] [Google Scholar]
- 15.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Proc Natl Acad Sci USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M. Proc Natl Acad Sci USA. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandir AS, Przedborski S, Jackson-Lewis V, Wang ZQ, Simbulan-Rosenthal CM, Smulson ME, Hoffman BE, Guastella DB, Dawson VL, Dawson TM. Proc Natl Acad Sci USA. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. J Neurosci. 2004;24:10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Zulueta M, Ensz LM, Mukhina G, Lebovitz RM, Zwacka RM, Engelhardt JF, Oberley LW, Dawson VL, Dawson TM. J Neurosci. 1998;18:2040–2055. doi: 10.1523/JNEUROSCI.18-06-02040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zelphati O, Wang Y, Kitada S, Reed JC, Felgner PL, Corbeil J. J Biol Chem. 2001;276:35103–35110. doi: 10.1074/jbc.M104920200. [DOI] [PubMed] [Google Scholar]
- 22.Alvarez-Gonzalez R, Jacobson MK. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 23.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 24.Winstall E, Affar EB, Shah R, Bourassa S, Scovassi AI, Poirier GG. Exp Cell Res. 1999;246:395–398. doi: 10.1006/excr.1998.4321. [DOI] [PubMed] [Google Scholar]
- 25.Miwa M, Tanaka M, Matsushima T, Sugimura T. J Biol Chem. 1974;249:3475–3482. [PubMed] [Google Scholar]
- 26.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 27.Patel CN, Koh DW, Jacobson MK, Oliveira MA. Biochem J. 2005;388:493–500. doi: 10.1042/BJ20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto S, Xue R, Sugo N, Sawada M, Blizzard KK, Poitras MF, Johns DC, Dawson TM, Dawson VL, Crain BJ, et al. Stroke. 2002;33:1101–1106. doi: 10.1161/01.str.0000014203.65693.1e. [DOI] [PubMed] [Google Scholar]
- 29.Dawson VL, Dawson TM, Bartley DA, Uhl GR, Snyder SH. J Neurosci. 1993;13:2651–2661. doi: 10.1523/JNEUROSCI.13-06-02651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Affar EB, Duriez PJ, Shah RG, Winstall E, Germain M, Boucher C, Bourassa S, Kirkland JB, Poirier GG. Biochim Biophys Acta. 1999;1428:137–146. doi: 10.1016/s0304-4165(99)00054-9. [DOI] [PubMed] [Google Scholar]
- 31.Haince JF, Poirier GG, Kirkland JB. In: Current Protocols in Cell Biology. Bonifacion JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada K, editors. Indianapolis: Wiley; 2005. Unit 18.7. [Google Scholar]
- 32.Cortez CM, Costa WS, Babinski MA, Chagas MA. Int J Morphol. 2003;21:221–226. [Google Scholar]
- 33.Longa EZ, Weinstein PR, Carlson S, Cummins R. Stroke. 1989;20:84–91. doi: 10.1161/01.str.20.1.84. [DOI] [PubMed] [Google Scholar]
- 34.Belayev L, Khoutorova L, Zhao W, Vigdorchik A, Belayev A, Busto R, Magal E, Ginsberg MD. Stroke. 2005;36:1071–1076. doi: 10.1161/01.STR.0000160753.36093.da. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.