Abstract
The TGF-β signaling pathway is a key organizer of injury and immune responses, and recent studies suggest it fulfills critical roles in CNS function and maintenance. TGF-β receptor activation results in phosphorylation of Smad proteins, which subsequently translocate to the nucleus to regulate gene transcription by binding to Smad binding elements (SBE). Using SBE-luciferase reporter mice, we recently discovered that the brain has the highest Smad baseline activity of any major organ in the mouse, and we now demonstrate that this signal is primarily localized to pyramidal neurons of the hippocampus. In vivo excitatory stimulation with kainic acid (KA) resulted in an increase in luciferase activity and phosphorylated Smad2 (Smad2P), and nuclear translocation of Smad2P in hippocampal CA3 neurons correlated significantly with luciferase activity. Although this activation was most prominent at 24 h after KA administration in neurons, Smad2P immunoreactivity gradually increased in astrocytes and microglial cells at 3 and 5 days, consistent with reactive gliosis. Bioluminescence measured over the skull in living mice peaked at 12–72 h and correlated with the extent of microglial activation and pathological markers of neurodegeneration 5 days after injury. Treatment with the glutamate receptor antagonist MK-801 strongly reduced bioluminescence and pathology. These results show that Smad2 signaling is a sensitive marker of neuronal activation and CNS injury that can be used to monitor KA-induced neuronal degeneration. This and related mouse models may provide valuable tools to study mechanisms and treatments for neurodegeneration.
Keywords: TGF-β, kainic acid, gliosis
The TGF-β superfamily of proteins, including TGF-βs, activins, and bone morphogenic proteins (BMPs) control cellular processes ranging from patterning and differentiation to proliferation and apoptosis, and they have been implicated in brain injury and neurodegeneration (1, 2). TGF-βs activate the TGF-β signaling pathway through a high-affinity transmembrane receptor complex consisting of the TGF-β type I (ALK5) and type II serine/threonine kinase receptor subunits (2, 3). TGF-β binding leads to phosphorylation of ALK5 and recruitment and phosphorylation of receptor-regulated Smad2 or Smad3. Once phosphorylated, these Smads associate with Smad4 and translocate into the nucleus where they can bind to Smad binding elements (SBE) in the DNA to regulate gene transcription (3). Activins, nodal, growth and differentiation factor (GDF)-8/myostatin, GDF-9, and GDF-11 signal also via Smad2/3 and Smad4 proteins by engaging activin receptors or ALK5 (3–7). In contrast, BMPs recruit Smad1, Smad5, and Smad8 in combination with Smad4 after binding to BMP type I and type II receptors (2, 3). Because of their distribution and expression patterns, TGF-β and activins are likely the main activators of Smad signaling in the brain.
Three TGF-β isoforms (TGF-β1, TGF-β2, and TGF-β3) have been described in the nervous system. Under normal conditions, TGF-β1 appears restricted to meningeal cells, choroid plexus epithelial cells, and glial cells, whereas TGF-β2 and TGF-β3 are expressed in both glia and neurons (8). TGF-β1 plays a central role in the response to brain injury and has been implicated in a number of disorders of the CNS, including stroke (9), Parkinson's disease (10), Alzheimer's disease (11), and brain tumors (12). Increased TGF-β1 expression is found in neurons after ischemia (13) and in Alzheimer's disease (14). Up-regulation of activin also was observed after stroke-induced (15) and kainic acid (KA)-induced excitotoxic injury (16), and activin was found to be of neuronal origin. Mice deficient in TGF-β1 show increased neuronal apoptosis and susceptibility to degeneration (17). Together, these studies underline the importance of TGF-βs and activin in normal CNS function and in response to disease and injury. However, it is unknown which cells in the brain respond to TGF-β signals and how signaling levels and patterns change in response to injury over time. Such knowledge is critical for our understanding and possible therapeutic targeting of this pathway in disease.
To monitor activation of the TGF-β signaling pathway in vivo, we recently have developed transgenic reporter mice, which contain a luciferase reporter gene fused to a promoter with multiple SBE (18). These mice showed highest basal Smad-dependent signaling in the brain, but we did not determine the cellular origin of this signal. Using bioluminescence imaging, we also demonstrated prominent reporter gene activation in the gut and the brain after treating the mice with lipopolysaccharide (LPS), and we were able to monitor gene activation in living mice over time (18). Bioluminescence has been used increasingly to monitor and quantify gene activity repeatedly in the same animal and has been used to study disease progression in peripheral organs with great success (19, 20). Bioluminescence imaging is quantitative and can faithfully report gene activation if appropriate fusion gene constructs are used (21, 22). Therefore, monitoring activation of an injury-responsive gene by bioluminescence imaging could provide valuable information about injury severity and disease progression.
Here, we demonstrate that Smad signaling is constitutively activated in hippocampal pyramidal neurons in mice and that neurons prominently increase Smad2 signaling after excitatory stimulation with KA. Three days after administration of this excitotoxin, glial cells showed increasingly activated Smad2 signaling. Bioluminescence imaging in living SBE-luc mice correlated with microglial activation and neurodegeneration after excitotoxic injury.
Results
Basal Smad2 Signaling in CNS Neurons.
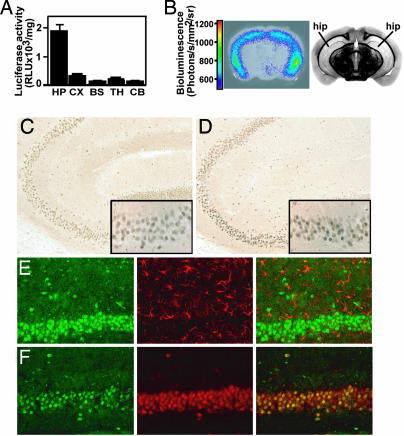
Studies in reporter mice for the TGF-β signaling pathway showed that basal signaling is highest in the brain of all major organs (18), consistent with an important homeostatic function of this pathway in the CNS. To localize this signaling activity, we dissected brains of 2-month-old SBE-luc mice (line T9-7F) into different brain regions. Biochemical measurements of luciferase activity showed by far the highest signal in the hippocampus and lower signals in the neocortex and thalamus (Fig. 1A). In agreement with these biochemical measurements, bioluminescence imaging of coronal brain slices of SBE-luc mice showed that photons were emitted mainly from the hippocampus, in particular from the subiculum and the distal portion of the CA1 subfield (Fig. 1B and Fig. 5, which is published as supporting information on the PNAS web site). Similar results were obtained in 7-day-old mice (line T9-55F), which had >10 times higher basal luciferase activity in the hippocampus compared with other brain regions (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Basal level of the luciferase reporter gene and Smad signaling in SBE-luc mice (T9-7F) and activation by excitotoxic injury. (A) Luciferase activity in brain regions of SBE-luc mice (n = 5). HP, hippocampus; CX, cortex; BS, brainstem; TH, thalamus; CB, cerebellum. (B) Bioluminescence of a 2-mm-thick coronal brain slice rapidly isolated from a SBE-luc mouse. The slice (Left) was bathed in tissue culture medium and luciferin, and bioluminescence was measured. The image from the brain atlas (Right) is shown to illustrate the localization and shape of hippocampus in the slice. (C and D) Sagittal vibratome sections from wild-type mice treated with saline (C) or KA (D) were stained with an antibody against Smad2P. Insets show CA3 region at higher magnification. (E and F) Sections from KA-treated mice were labeled for Smad2P (green) in combination with GFAP (red, E) or NeuN (red, F) by using fluorescently labeled secondary antibodies. The merged images are shown at Right.
To determine in which CNS cells the TGF-β signaling pathway is activated, we used two different antibodies raised against the activated, phosphorylated form of Smad2 (Smad2P) (23, 24). Interestingly, Smad2P was localized predominantly to neurons and, in agreement with the biochemical and bioluminescent data, to hippocampal pyramidal neurons (Fig. 1C and data not shown). Scattered large pyramidal neurons in layer 4 of the neocortex also showed Smad2P immunoreactivity, but little or no signal was detected in astrocytes or microglia throughout the brain (data not shown). In contrast, in vivo administration of the glutamate receptor agonist KA resulted in a widespread increase in Smad2P, with a translocation from the cytoplasm to the nucleus, as shown in CA3 pyramidal neurons (Fig. 1D). Again, the signal was restricted largely to neurons (Fig. 1 E and F). Relative intensity of Smad2P immunostaining in CA3 neurons of the hippocampus of KA-treated SBE-luc mice correlated positively with luciferase activity measured in hippocampal homogenates from the opposite hemibrain (R = 0.85, P = 0.002, n = 10 mice, linear regression analysis). We also tested several antibodies from commercial or academic sources against Smad3P, but they all cross-reacted with Smad2P or were otherwise nonspecific. Our results demonstrate that Smad2 signaling is activated prominently in the brain in response to neuronal injury, notably in hippocampal neurons that are known to be susceptible to excitotoxic injury (25). The results also validate SBE-luc mice as a model to study Smad signaling by showing that luciferase activity correlates with endogenous Smad2 phosphorylation.
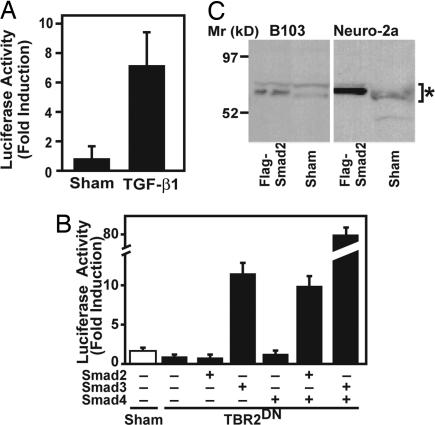
To confirm that neurons can indeed respond directly to Smad2 and TGF-β signals, we cultured primary neurons from brains of SBE-luc mice. Treatment with recombinant TGF-β1 resulted in a clear increase in luciferase activity (Fig. 2A). In addition, transient transfection of Neuro-2a mouse neuroblastoma cells showed that Smad2 in combination with Smad4, although less potent than Smad3/4, was able to induce activation of a SBE-luciferase reporter gene (Fig. 2B). Western blot analysis also demonstrated that neuroblastoma cells express Smad2 (Fig. 2C). Thus neurons can be stimulated directly with TGF-βs to activate Smad2.
Fig. 2.
Specificity of SBE-luc reporter gene. (A) Primary neurons cultured from postnatal day 2 forebrains of SBE-luc mice (line T9-55F) were stimulated with recombinant TGF-β1 (1 ng/ml) or buffer as a control. Bars are mean ± SEM from one of three experiments. (B) Neuro-2a mouse neuroblastoma cells were transiently transfected with SBE-luc reporter plasmid and various combinations of CMV-Flag-Smad plasmids. CMV-TBR2DN was transfected to suppress the endogenous high basal TGF-β signaling. Bars are mean ± SEM from one of three independent experiments. (C) Immunoblot of cell lysates from two neuronal cell lines with an antibody against Smad2 shows expression of Smad2 (Sham). Cells transiently transfected with Flag-Smad2 were included as positive controls. The two different size bands of endogenous or Flag-Smad2 (*) may represent alternatively spliced form and/or posttranslational modifications.
Glial Activation of Smad2 Signaling in Response to Neuronal Injury.
Although Smad2 signaling in untreated mice or 24 h after KA treatment was predominantly neuronal (Fig. 1 C–F), we wanted to determine whether this expression pattern changes in response to neuronal injury. KA causes excitotoxicity by inducing an excessive activation of glutamate receptors and subsequent Ca2+ influx in neurons. The resulting neuronal injury causes an inflammatory response and oxidative stress that amplifies neuronal degeneration and death (26). Excitotoxicity has been postulated to be responsible not only for neuronal loss associated with seizures, traumatic and ischemic brain injury, and hypoxia (27) but also for neuronal damage in Alzheimer's disease and other neurodegenerative disorders (25).
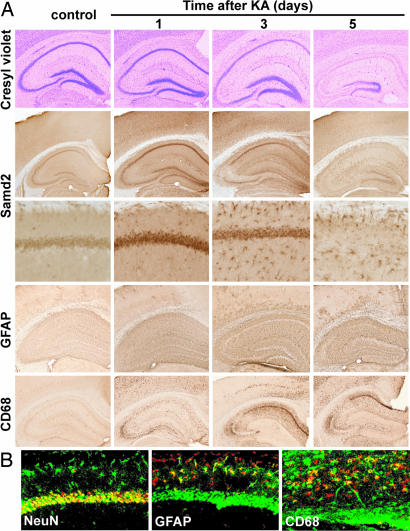
Cresyl violet staining confirmed the progressive degeneration of neurons 1–5 days after KA administration in CA pyramidal neurons of the hippocampus, hilar neurons, and, eventually, granule cells of the dentate gyrus (Fig. 3A). Along with this degeneration, glial cells are activated. Although microglia show some activation at 1 day around CA3 pyramidal neurons, more prominent activation of microglia and astrocytes is detected at 3 and 5 days (Fig. 3 A and B). Interestingly, activated astrocytes and, to a lesser extent, microglia show prominent Smad2P immunoreactivity at these later stages. At the same time, the neuronal signal, which peaked at 24 h, decreases in intensity and, because of neuronal loss, in abundance (Fig. 3 A and B). Together, the Smad2P signal appears to correlate with the relative gliosis and neuronal damage over time.
Fig. 3.
Neuronal injury, glial activation, and activation of Smad signaling after excitotoxic brain injury. SBE-luc mice (T9-7F) were lesioned with KA (20 mg/kg, s.c.) and killed 1, 3, and 5 days later. (A) Time course of KA-induced neuronal injury (cresyl violet staining), activation of Smad signaling (Smad2P immunostaining), astroglial activation (GFAP immunostaining), and microglial activation (CD68 immunostaining). (B) Confocal microscope images of double immunofluorescence with antibodies against Smad2P (24) (green) and cell type-specific markers NeuN, CD68, or GFAP (red). Smad2P-expressing cells appear yellow after superimposition.
Noninvasive Imaging of Neuronal Injury in Living Mice and Correlation with Pathology.
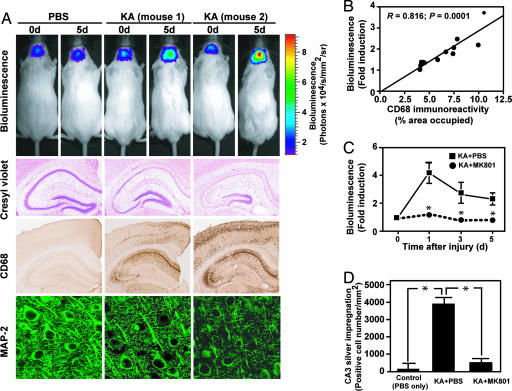
To test whether Smad signaling could be used as a surrogate marker to estimate gliosis and neuronal injury in this model, we measured bioluminescence in living SBE-luc mice treated with KA. An i.p. injection of luciferin into SBE-luc mice resulted in detectable emission of photons over the skull, and this signal increased in KA-treated mice (Fig. 4A). The signal, which originated almost exclusively from brain tissue (Fig. 7, which is published as supporting information on the PNAS web site), increased severalfold 12 h after KA treatment and remained elevated until it decreased 4–5 days later (Fig. 8, which is published as supporting information on the PNAS web site). Neuropathological analysis of mice 5 days after KA treatment showed consistent loss of hippocampal CA neurons and gliosis although considerable differences in the extent of damage were observed between individual mice (Fig. 4A, two mice with different extent of damage and corresponding luciferase activity are shown). Notably, the relative increase over baseline in bioluminescence signals at 5 days correlated strongly with postmortem microgliosis (Fig. 4B and Fig. 9, which is published as supporting information on the PNAS web site), consistent with a relationship between Smad2P immunostaining and microglial activation in this model (Fig. 3A). In a second independent experiment, we lesioned mice from line T9-55 with KA and found a similar, highly significant correlation between microglial activation and reporter gene activation measured as bioluminescence (R = 0.90, P < 0.0001, n = 11 mice, linear regression analysis; Table 2, which is published as supporting information on the PNAS web site).
Fig. 4.
KA-induced neuropathology and treatment with MK801. (A and B) SBE-luc mice (T9-7F) were lesioned with KA (20 mg/kg, s.c.) and killed 5 days later. Bioluminescence was recorded in living mice injected with luciferin (150 mg/kg) before they were killed and is expressed as fold induction over baseline measured 1 day before KA administration. Neuronal injury and degeneration was detected by cresyl violet staining and immunohistochemical staining with an antibody against MAP-2. Microglial activation was assessed by using CD68 immunostaining. (A) Images from representative mice injected with PBS (Left) or KA (Center and Right) show mice with different extent of injury and reporter gene activity. (B) Correlation between bioluminescence and hippocampal microglial activation (assessed by CD68 immunoreactivity). (C and D) Pretreatment with MK-801 reduced bioluminescence (C) and neuronal injury (D) in KA-treated mice. SBE-luc mice (T9-7F) were lesioned with KA (30 mg/kg, s.c.), and bioluminescence was recorded at the indicated times in each mouse. MK-801 (1 mg/kg, i.p.) or PBS was administered 1 h before KA. Bars are mean ± SEM (n = 3). ∗, P < 0.01 compared with KA + PBS by t test (C) or by ANOVA and Dunnet's test (D).
Similarly, loss of hippocampal neurons measured by cresyl violet staining (Fig. 4A and Fig. 10, which is published as supporting information on the PNAS web site) or general neuronal damage measured by silver impregnation correlated strongly with bioluminescence (Table 1 and Fig. 11, which is published as supporting information on the PNAS web site). In addition, several other markers of neuronal and synapto-dendritic integrity, including NeuN (Fig. 12, which is published as supporting information on the PNAS web site), MAP-2 (Fig. 4A), synaptophysin, and calbindin, which frequently are used to assess neurodegeneration in a number of diseases and animal models (28–31), showed highly significant inverse correlations with bioluminescence at 5 days (Table 1), indicating that Smad signaling is a sensitive marker of neuronal injury and damage. Similar correlations between bioluminescence and neurodegeneration also were obtained in line T9-55F (Table 2).
Table 1.
Correlation between bioluminescence (fold induction) and markers of neuronal injury
| Markers of neuronal injury | Hippocampus |
Cortex |
||
|---|---|---|---|---|
| R | P | R | P | |
| Cresyl violet (area) | 0.698 | 0.009 | NA | |
| Silver staining | ||||
| CA3 | 0.731 | 0.005 | NA | |
| Cortex | NA | 0.679 | 0.013 | |
| Synaptophysin | −0.701 | 0.009 | −0.699 | 0.009 |
| MAP-2 | −0.734 | 0.005 | −0.755 | 0.003 |
| NeuN | −0.771 | 0.002 | −0.791 | 0.001 |
| Calbindin | −0.797 | 0.001 | ND | |
| Microgliosis (CD68) | 0.816 | 0.0001 | 0.878 | <0.0001 |
Correlation between bioluminescence and markers of neuronal injury and microglial activation. SBE-luc mice (T9-7F, n = 11) lesioned with KA (20 mg/kg, s.c.) were killed 5 days after treatment. Bioluminescence was recorded in living mice injected with luciferin (150 mg/kg) before they were killed and is expressed as fold induction over baseline measured 1 day before KA administration. Neuronal injury was measured by quantifying cresyl violet staining, silver impregnation, and immunolabeling for synaptophysin, MAP-2, NeuN, and calbindin. Microglial activation was quantified from sections stained with CD68 antibody. These markers were quantified separately in hippocampus and cortex by using image analysis software, and the correlation between these markers and bioluminescence was assessed by Pearson correlation analyses. R, correlation coefficient; NA, not applicable; ND, not done.
To demonstrate the specificity and potential use of Smad-dependent bioluminescence imaging to monitor neuronal injury, we treated SBE-luc mice with MK-801, an anticonvulsant drug that antagonizes NMDA receptor signaling and the effects of KA (32). MK-801 treatment completely prevented KA-induced activation of the SBE-luc gene and induction of bioluminescence (Fig. 4C), and neuropathological analysis revealed minimal neurodegeneration (Fig. 4D) and microgliosis (data not shown).
Discussion
The present study demonstrates that neurons constitutively signal through Smad2 proteins in the hippocampus and that Smad2-dependent signaling increases after excitatory stimulation first in neurons and subsequently in glial cells. This signaling can be imaged noninvasively in living SBE-luc reporter mice in the form of bioluminescence emitted from the skull. Importantly, bioluminescence signals from living SBE-luc reporter mice treated with the excitotoxin KA correlated with several markers of neuronal injury and degeneration as well as with microglial activation. This finding indicates that Smad signaling can be used as a surrogate marker to quantify CNS injury in this model.
TGF-β1 and activin previously have been reported to be expressed by neurons and glial cells and increased in response to a number of CNS injuries (9). We show here that adult hippocampal neurons maintain constitutive activation of Smad2 (Smad2P), a downstream mediator of the TGF-β signaling pathway (Fig. 1). In addition, luciferase activity in SBE-luc mice was by far highest in the hippocampus of all brain regions. These studies indicate that there is persistent signaling of TGF-βs and/or activin in the hippocampus and implicate Smad2 in normal neuronal function. These data are consistent with findings in Smad2 knockout mice (heterozygous), which have spatial learning and memory deficits in the hippocampal-dependent version of the water maze, but their learning and memory in tasks that depend primary on amygdala function are normal.¶
Neuronal Smad2 also was found to be prominently activated in response to stimulation with KA (Fig. 1), a glutamate receptor agonist that causes excitotoxicity. We cannot conclude from our studies whether this increase in signaling is a protective response or contributes to cell death, but earlier studies in TGF-β1-deficient mice or in primary neurons derived from such mice suggest that TGF-β signaling and activation of Smad2 is neuroprotective (17). This does not rule out that TGF-β may signal via other pathways and regulate neuronal activity or survival. For example, KA can induce up-regulation of ALK1, an alternative TGF-β type I receptor used by endothelial cells (33) and neurons (34), and signal via Smad1 (34). It also is interesting that Smad2 activation is delayed in glial cells and not readily detectable 24 h after KA administration. It is possible, therefore, that injured neurons release TGF-βs or activin, which in turn activate Smad signaling in glia.
Activation of luciferase in SBE-luc mice after KA correlated with Smad2P translocation to the nucleus, underlining that this reporter indeed measures Smad signaling. Furthermore, overall Smad2 activation 3 or 5 days after KA administration appeared to correlate with glial cell activation (Fig. 3). Therefore, we tested whether Smad signaling and activation of luciferase in reporter mice could be measured to predict neuronal injury and degeneration. Indeed, bioluminescence imaging in living mice showed that reporter gene activation correlates strongly with several markers of neuronal injury and degeneration as well as with microglial activation (Fig. 4 and 9–12 and Tables 1 and 2). Treatment of mice with MK-801 prevented activation of the reporter gene in response to KA, consistent with a lack of neuronal injury in the postmortem analysis of these mice (Fig. 4). We conclude that Smad2 signaling is a sensitive indicator of CNS injury in this model and can be used to monitor such injury and test potential neuroprotective drugs.
Bioluminescence imaging has been used recently for the study of gene expression in transgenic mice that express luciferase reporter genes under control of various promoters or transcription factor binding sites (35–38). In the brain, bioluminescence has been used mainly to track and quantify luciferase-tagged tumor cells (39–42) or to trace migration of neural progenitor cells (43, 44). One study used glial fibrillary acidic protein (GFAP)-luciferase transgenic mice to demonstrate activation of the reporter after injury, but no correlation with neuronal injury or degeneration was demonstrated (38). Reporter mice such as SBE-luc mice should be useful to carry out simple pharmacology studies, dose-testing, and assessment of blood–brain–barrier permeability. Notably, relatively small numbers of animals are sufficient to detect even small drug effects over time because each mouse serves as its own control.
In conclusion, we show here that the TGF-β signaling pathway, a key signaling pathway involved in multiple biological processes, is constitutively active in neurons and is activated after neuronal excitation and injury in vivo. Transgenic SBE-luc reporter mice for this pathway were used to study signaling and monitor pathway activation in living mice, and we demonstrate they can be used to rapidly test pharmacological compounds. Similar reporter mouse models could be engineered to respond to different cellular pathways and perhaps be used to monitor neural activity and other more complex physiological and behavioral responses.
Methods
Mice and Injury Models.
Transgenic mouse lines T9-7F or T9-55F on the FVB/N genetic background harboring a SBE-luc transgene have been described (18). This transgene consists of 12 SBE repeats fused to a herpes simplex virus/thymidine kinase minimal promoter upstream of firefly luciferase followed by a simian virus 40 late polyadenylation signal (18). The mice are now available at The Jackson Laboratory (stock no. 005999; Bar Harbor, ME). Heterozygous, 2- to 3-month-old mice with detectable luciferase activity in tail biopsies [“responder” mice (18)] were used in all experiments. For excitotoxin-induced injury, KA (Tocris, Ellisville, MO) was dissolved in PBS and injected i.p. (25 mg/kg) or s.c. (20 mg/kg) to induce neurodegeneration. Seizure activity was scored from 0 to 5, with 0 showing no behavioral changes and 5 showing constant rearing and falling (45). All KA-injected mice reached at least stage 3. For pretreatment with anticonvulsant, MK-801 (dizocilpine, 1 mg/kg; Sigma-Aldrich, St Louis, MO) was prepared in PBS and administrated i.p. 1 h before KA injection. Control animals in all experiments received either PBS or saline injections. All animal handling was performed in accordance with institutional guidelines and approved by the local Institutional Animal Care and Use Committee.
Cell Culture.
Neuronal cortical cultures were prepared from fetal mice at embryonic days 15–16 as described (46). Cells were cultured in DMEM supplemented with 2 mM glutamine, 5% horse serum, and 5% FBS. Cytarabine (Ara-C) was added 3 days after plating to inhibit nonneuronal cell proliferation. Five-day-old primary neurons were stimulated with recombinant TGF-β1 (R & D Systems, Minneapolis, MN). Cells were harvested 16 h later and lysed in 100 μl of 1× Cell Culture Lysis Reagent (Promega, Madison, WI), and luciferase activity was measured with a luciferase assay kit (Promega) by using a tube luminometer (Lumat LB 9507; Berthold Technologies, Oak Ridge, TN).
Mouse neuroblastoma cells Neuro-2a (CCL-131; ATCC, Manassas, VA) were cultured in Eagle's minimum essential medium with 2 mM glutamine, 1.5 g/liter sodium bicarbonate, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 10% FBS (Invitrogen, Carlsbad, CA). To differentiate Neuro-2a cells, cells were maintained in Neurobasal media with 2 mM glutamine and 1× N2 supplement (Invitrogen). Rat neuroblastoma cells B103 stably expressing wild-type human amyloid precursor protein and neomycin resistance gene (47) were cultured in DMEM with 2 mM glutamine and 10% FBS. Neuro-2a and B103 cells were transfected with the following plasmids: CMV-Flag-Smad2 (0.3 μg per well), CMV-Flag-Smad3 (0.3 μg per well), and CMV-Flag-Smad4 (0.1 μg per well; a kind gift from Rik Derynck, University of California, San Francisco, CA). In addition, Neuro-2a cells were cotransfected with SBE-Luc reporter plasmid (0.1 μg per well) and CMV-TBR2DN (dominant-negative TGF-β type 2 receptor, 0.3 μg per well) to suppress the endogenous high basal TGF-β signaling in these cells. Luciferase activity was measured 16 h later as described above.
Immunoblotting.
Differentiated Neuro-2a or B103 cells (1 × 106) were harvested and lysed in 200 μl of RIPA lysis buffer [500 mM Tris (pH 7.4), 150 mM NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, and complete protease inhibitors (Roche Basel, Switzerland)]. Cell lysates (20 μl) were mixed with 4× NuPage LDS loading buffer (Invitrogen) and loaded on a 3–12% SDS/polyacrylamide gradient gel and subsequently transferred onto a PVDF membrane. The blot was incubated with rabbit polyclonal antibodies (1:500) against Smad2 (23) and a horseradish peroxidase-conjugated donkey anti-rabbit IgG as secondary antibody (Amersham Pharmacia Biotech, Piscataway, NJ). Protein signals were detected by using an ECL kit (Amersham Pharmacia Biotech).
Tissue Preparations.
Mice were anesthetized with 400 mg/kg chloral hydrate (Sigma-Aldrich) and transcardially perfused with 0.9% saline. Brains were dissected, and one hemibrain was fixed for 24 h in 4% paraformaldehyde and cryoprotected in 30% sucrose. Serial coronal sections (40-μm) were cut with a freezing microtome (Leica, Allendale, NJ) and stored in cryoprotective medium. One set of sections, representing different levels of the hippocampus, was used for each staining. The other hemibrain was dissected into hippocampus, cortex, thalamus, brainstem, and cerebellum. Each subregion was weighed and lysed in 100–400 μl of 1× Cell Culture Lysis Reagent (Promega). Luciferase activities from tissue homogenates were measured as described above and normalized to weight.
Detection of Neurodegeneration and Microgliosis.
Brain sections were mounted on Superfrost plus slides (Fisher Scientific, Pittsburgh, PA) and stained with 0.02% cresyl violet (Sigma) (17). Degenerating neurons were detected by silver impregnation with the FD Neurosilver kit (FD NeuroTechnologies, Ellicott City, MD) according to the manufacturer's protocol. Immunohistochemistry was performed on free-floating sections following a standard procedure (48). The following primary antibodies were used: anti-Smad2P [1:200 (23) or 1:100 (24)]; anti-NeuN (1:1,000; Chemicon, Temecula, CA); anti-MAP-2 (1 μg/ml; Roche); anti-synaptophysin (1:800; Roche); anti-calbindin (1:2,500; Sigma); anti-GFAP (1:1,000; Dako, Carpinteria, CA); and anti-CD68 (1:50; Serotec, Raleigh, NC). Primary antibody staining was revealed by biotinylated secondary antibodies and the ABC kit (Vector Laboratories, Burlingame, CA) with diaminobenzidine (Sigma-Aldrich). For double immunolabeling, the Smad2P signal was amplified by tyramide signal amplification (TSA) kit (PerkinElmer Life Sciences, Boston, MA). For detailed experimental procedures and image analysis, see Supporting Methods, which is published as supporting information on the PNAS web site.
Bioluminescence Imaging.
Bioluminescence was detected with the In Vivo Imaging System (18) (IVIS; Xenogen, Alameda, CA). Mice were injected i.p. with 150 mg/kg d-luciferin (Xenogen) 10 min before imaging and anesthetized with isoflurane during imaging. Photons emitted from living mice were acquired as photons per s/cm2 per steridian (sr) by using LIVINGIMAGE software (Xenogen) and integrated over 5 min. For photon quantification, a region of interest was manually selected and kept constant within all experiments; the signal intensity was converted into photons per s/cm2 per sr. For longitudinal comparison of bioluminescence, baseline imaging was performed 24 h before KA was administered. Bioluminescence was expressed as fold induction over baseline levels. In addition, a background bioluminescence reading obtained in nontransgenic mice injected with luciferin was subtracted from all values. For brain slice imaging, coronal brain slices (2- to 3-mm thickness) were obtained by cutting brains in a mouse brain matrix (Harvard Apparatus, Holliston, MA). Slices were bathed in tissue culture medium containing luciferin (150 μg/ml) in 24-well plates and imaged immediately.
Statistical Analysis.
Data were expressed as mean ± SEM. Statistical analyses were performed with Prism 4.03 software (GraphPad, San Diego, CA). Correlation coefficients were calculated with Pearson correlation analyses. Means between two groups were compared with two-tailed, unpaired Student's t tests; comparisons of means from multiple groups with one control were analyzed with one-way ANOVA and Dunnet's post hoc test.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AG20603 and AG023708 (to T.W.-C.), the Veterans Administration Geriatric Research, Education, and Clinical Center and Mental Illness Research, Education, and Clinical Center services (T.W.-C.). A.H.L. was supported by a National Research Service Award.
Abbreviations
- SBE
Smad binding elements
- KA
kainic acid
- Smad2P
phosphorylated Smad2
- GFAP
glial fibrillary acidic protein.
Footnotes
¶Frankland, P. W., Wang, Y., Heyer, J., Kucherlapati, R., Silva, A. (2001) Soc. Neurosci. Abstr. 27:1980 (abstr.).
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Massagué J, Blain SW, Lo RS. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 2.Dennler S, Goumans MJ, ten Dijke P. J Leukoc Biol. 2002;71:731–740. [PubMed] [Google Scholar]
- 3.Shi Y, Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 4.Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH. Genes Dev. 2001;15:2010–2022. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh SP, Yeo CY, Lee Y, Schrewe H, Whitman M, Li E. Genes Dev. 2002;16:2749–2754. doi: 10.1101/gad.1021802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Mol Cell Biol. 2003;23:7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazerbourg S, Klein C, Roh J, Kaivo-Oja N, Mottershead DG, Korchynskyi O, Ritvos O, Hsueh AJ. Mol Endocrinol. 2004;18:653–665. doi: 10.1210/me.2003-0393. [DOI] [PubMed] [Google Scholar]
- 8.Unsicker K, Flanders KC, Cissel DS, Lafyatis R, Sporn MB. Neuroscience. 1991;44:613–625. doi: 10.1016/0306-4522(91)90082-y. [DOI] [PubMed] [Google Scholar]
- 9.Kim JS, Yoon SS, Kim YH, Ryu JS. Stroke. 1996;27:1553–1557. doi: 10.1161/01.str.27.9.1553. [DOI] [PubMed] [Google Scholar]
- 10.Mogi M, Harada M, Kondo T, Narabayashi H, Riederer P, Nagatsu T. Neurosci Lett. 1995;193:129–132. doi: 10.1016/0304-3940(95)11686-q. [DOI] [PubMed] [Google Scholar]
- 11.van der Wal EA, Goḿez-Pinilla F, Cotman CW. NeuroReport. 1993;4:69–72. doi: 10.1097/00001756-199301000-00018. [DOI] [PubMed] [Google Scholar]
- 12.Platten M, Wick W, Weller M. Microsc Res Tech. 2001;52:401–410. doi: 10.1002/1097-0029(20010215)52:4<401::AID-JEMT1025>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Knuckey NW, Finch P, Palm DE, Primiano MJ, Johanson CE, Flanders KC, Thompson NL. Mol Brain Res. 1996;40:1–14. doi: 10.1016/0169-328x(96)00016-2. [DOI] [PubMed] [Google Scholar]
- 14.Ueberham U, Ueberham E, Gruschka H, Arendt T. Neuroscience. 2003;116:1–6. doi: 10.1016/s0306-4522(02)00670-x. [DOI] [PubMed] [Google Scholar]
- 15.Bottner M, Dubal DB, Rau SW, Suzuki S, Wise PM. J Neuroendocrinol. 2006;18:97–103. doi: 10.1111/j.1365-2826.2005.01384.x. [DOI] [PubMed] [Google Scholar]
- 16.Tretter YP, Hertel M, Munz B, Bruggeencate GT, Werner S, Alzheimer C. Nat Med. 2000;6:812–815. doi: 10.1038/77548. [DOI] [PubMed] [Google Scholar]
- 17.Brionne TC, Tesseur I, Masliah E, Wyss-Coray T. Neuron. 2003;40:1133–1145. doi: 10.1016/s0896-6273(03)00766-9. [DOI] [PubMed] [Google Scholar]
- 18.Lin AH, Luo J, Mondshein LH, Ten Dijke P, Vivien D, Contag CH, Wyss-Coray T. J Immunol. 2005;175:547–554. doi: 10.4049/jimmunol.175.1.547. [DOI] [PubMed] [Google Scholar]
- 19.Weissleder R, Ntziachristos V. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 20.Welsh DK, Kay SA. Curr Opin Biotechnol. 2005;16:73–78. doi: 10.1016/j.copbio.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Greer LF III, Szalay AA. Luminescence. 2002;17:43–74. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- 22.Contag CH, Bachmann MH. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 23.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. EMBO J. 2002;21:1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan W, Vellucci VF, Reiss M. Oncol Res. 2000;12:157–167. doi: 10.3727/096504001108747639. [DOI] [PubMed] [Google Scholar]
- 25.Rothman SM, Olney JW. Trends Neurosci. 1995;18:57–58. doi: 10.1016/0166-2236(95)93869-y. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Yu S, Simonyi A, Sun GY, Sun AY. Mol Neurobiol. 2005;31:3–16. doi: 10.1385/MN:31:1-3:003. [DOI] [PubMed] [Google Scholar]
- 27.Choi DW. J Neurobiol. 1992;23:1261–1276. doi: 10.1002/neu.480230915. [DOI] [PubMed] [Google Scholar]
- 28.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Ann Neurol. 1991;30:572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 29.Masliah E, Achim CL, Ge N, DeTeresa R, Terry RD, Wiley CA. Ann Neurol. 1992;32:321–329. doi: 10.1002/ana.410320304. [DOI] [PubMed] [Google Scholar]
- 30.Buttini M, Orth B, Bellosta S, Akeefe H, Pitas RE, Wyss-Coray T, Mucke L, Mahley RW. J Neurosci. 1999;19:4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palop JJ, Jones B, Kekonius L, Chin J, Yu GQ, Raber J, Masliah E, Mucke L. Proc Natl Acad Sci USA. 2003;100:9572–9577. doi: 10.1073/pnas.1133381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zucchini S, Buzzi A, Bergamaschi M, Pietra C, Villetti G, Simonato M. NeuroReport. 2002;13:2071–2074. doi: 10.1097/00001756-200211150-00016. [DOI] [PubMed] [Google Scholar]
- 33.Goumans MJ, Valdimarsdottir G, Itoh S, Lebrin F, Larsson J, Mummery C, Karlsson S, ten Dijke P. Mol Cell. 2003;12:817–828. doi: 10.1016/s1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- 34.Konig HG, Kogel D, Rami A, Prehn JH. J Cell Biol. 2005;168:1077–1086. doi: 10.1083/jcb.200407027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Contag CH, Spilman SD, Contag PR, Oshiro M, Eames B, Dennery P, Stevenson DK, Benaron DA. Photochem Photobiol. 1997;66:523–531. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 36.Carlsen H, Moskaug JO, Fromm SH, Blomhoff R. J Immunol. 2002;168:1441–1446. doi: 10.4049/jimmunol.168.3.1441. [DOI] [PubMed] [Google Scholar]
- 37.Sadikot RT, Han W, Everhart MB, Zoia O, Peebles RS, Jansen ED, Yull FE, Christman JW, Blackwell TS. J Immunol. 2003;170:1091–1098. doi: 10.4049/jimmunol.170.2.1091. [DOI] [PubMed] [Google Scholar]
- 38.Zhu L, Ramboz S, Hewitt D, Boring L, Grass DS, Purchio AF. Neurosci Lett. 2004;367:210–212. doi: 10.1016/j.neulet.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Burgos JS, Rosol M, Moats RA, Khankaldyyan V, Kohn DB, Nelson MD, Jr, Laug WE. BioTechniques. 2003;34:1184–1188. doi: 10.2144/03346st01. [DOI] [PubMed] [Google Scholar]
- 40.Rehemtulla A, Hall DE, Stegman LD, Prasad U, Chen G, Bhojani MS, Chenevert TL, Ross BD. Mol Imaging. 2002;1:43–55. doi: 10.1162/15353500200200005. [DOI] [PubMed] [Google Scholar]
- 41.Soling A, Theiss C, Jungmichel S, Rainov NG. Genet Vaccines Ther. 2004;2:7. doi: 10.1186/1479-0556-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhrbom L, Nerio E, Holland EC. Nat Med. 2004;10:1257–1260. doi: 10.1038/nm1120. [DOI] [PubMed] [Google Scholar]
- 43.Tang Y, Shah K, Messerli SM, Snyder E, Breakefield X, Weissleder R. Hum Gene Ther. 2003;14:1247–1254. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- 44.Kim DE, Schellingerhout D, Ishii K, Shah K, Weissleder R. Stroke. 2004;35:952–957. doi: 10.1161/01.STR.0000120308.21946.5D. [DOI] [PubMed] [Google Scholar]
- 45.Schauwecker PE, Steward O. Proc Natl Acad Sci USA. 1997;94:4103–4108. doi: 10.1073/pnas.94.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buisson A, Nicole O, Docagne F, Sartelet H, MacKenzie ET, Vivien D. FASEB J. 1998;12:1683–1691. [PubMed] [Google Scholar]
- 47.Xu X, Yang D, Wyss-Coray T, Yan J, Gan L, Sun Y, Mucke L. Proc Natl Acad Sci USA. 1999;96:7547–7552. doi: 10.1073/pnas.96.13.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyss-Coray T, Lin C, Yan F, Yu G, Rohde M, McConlogue L, Masliah E, Mucke L. Nat Med. 2001;7:612–618. doi: 10.1038/87945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.