Abstract
To understand the mechanisms underlying mutagenesis in eukaryotes better, we have cloned mouse and human homologs of the Escherichia coli dinB gene. E. coli dinB encodes DNA polymerase IV and greatly increases spontaneous mutations when overexpressed. The mouse and human DinB1 amino acid sequences share significant identity with E. coli DinB, including distinct motifs implicated in catalysis, suggesting conservation of the polymerase function. These proteins are members of a large superfamily of DNA damage-bypass replication proteins, including the E. coli proteins UmuC and DinB and the Saccharomyces cerevisiae proteins Rev1 and Rad30. In a phylogenetic tree, the mouse and human DinB1 proteins specifically group with E. coli DinB, suggesting a mitochondrial origin for these genes. The human DINB1 gene is localized to chromosome 5q13 and is widely expressed.
In Escherichia coli, mutagenesis associated with exposure to DNA-damaging agents requires a specialized system (the SOS system), which processes the damage in an error-prone fashion, resulting in mutations (1). Recent in vitro studies with purified reconstituted systems have shown that E. coli UmuC protein, in conjunction with UmuD′ protein (both of which are encoded by SOS-regulated genes; ref. 1), single-strand binding protein, and activated RecA protein, can facilitate error-prone bypass of DNA lesions by DNA polymerase III holoenzyme (2, 3). The dinB gene of E. coli (sometimes referred to as dinP; ref. 4) is also regulated by the SOS system and is required for untargeted (spontaneous) mutations in phage λ when infected cells are exposed to UV radiation (5). Additionally, overexpression of the cloned dinB gene in unirradiated E. coli cells carrying plasmids dramatically increases the mutational burden in the plasmid DNA (6). Recently, E. coli DinB protein has been purified and shown to have a specialized DNA polymerase activity (7).
E. coli
DinB protein is homologous to an uncharacterized protein from Caenorhabditis elegans (F22B7.6), the Saccharomyces cerevisiae Rev1 protein, and E. coli UmuC protein (4). Like UmuC protein, Rev1 is involved in DNA damage-induced mutagenesis in yeast (8). Rev1 protein has been shown to possess an unusual type of DNA polymerase activity that efficiently inserts dCMP residues across from sites of base loss in a template/primer-dependent reaction; this enzyme has been called a deoxycytidyl transferase (9). More recently, the yeast Rad30 protein, which is also homologous to UmuC and DinB (10, 11), has been shown to be a DNA polymerase (DNA polymerase η) that accurately replicates thymine dimers in template DNA (12). A human homolog of Rad30 has properties very similar to that of yeast DNA polymerase η (13), and patients from the variant group of the cancer-prone hereditary disease xeroderma pigmentosum (XPV) have been shown to carry mutations in this homolog of RAD30 (14, 15). Collectively, these observations suggest that members of the UmuC/DinB superfamily are all replication-bypass DNA polymerases. However, these polymerases may differ in their fidelity and/or affinity for various types of damaged DNA.
Here, we report the cDNA and translated amino acid sequence of mouse and human homologs of the E. coli dinB gene. We have performed phylogenetic analysis of the UmuC/DinB superfamily and identified four distinct subfamilies. All members of the UmuC/DinB superfamily share predicted catalytic domains as well as helix–hairpin–helix (HhH) DNA-binding domains, consistent with the notion that they are DNA polymerases. Beyond these regions, the superfamily shows considerable diversity of domain architecture. In particular, the human and mouse DinB homologs contain a distinct version of a Zn-finger module that is found in several other enzymes implicated in DNA repair. The human gene (DINB1) maps to chromosome 5q13 and is expressed at low but varying levels in all tissues tested. Expression of DINB1 mRNA is highest in testis, and multiple transcripts are present in this tissue.
Materials and Methods
Cloning and Sequencing of the Mouse Dinb1 and Human DINB1 Genes.
Total RNA from mouse embryonic fibroblasts or mouse testis was used as a template for first-strand cDNA synthesis by using the Superscript Preamplification System (Life Technologies, Rockville, MD) according to the manufacturer’s directions. Degenerate primers were designed based on conserved sequences in the E. coli DinB and C. elegans F22B7.6 proteins. The degenerate primers capable of encoding C. elegans F22B7.6 amino acids 93–99 [YFAAVEM] and amino acids 289–296 [NKPNGQ(Y/F)V] were DPH1C (5′-CGA ATT CTA YTT YGC NGC IGT NGARAT G-3′) and DPH4NC (5′-CGG GAT CCA CRW AYT GIC CRT TIG GYT TRT T-3′), where Y = C/T, N = A/C/G/T, I = inosine, R = A/G, and W = A/T. PCRs were performed by using AmpliTaq polymerase and conditions recommended by the manufacturer (Perkin–Elmer). Touchdown PCR was performed with annealing at 60–51°C for 2 cycles and 50°C for 22 cycles. Amplification from mouse cDNA with these primers resulted in a product of 700 bp. This portion of the mouse Dinb1 gene was used to generate a random-primed probe for screening a mouse testis cDNA library and a human HeLa cell cDNA library. Two partial cDNA clones obtained from each library were sequenced. Multiple rounds of 5′ and 3′ rapid amplification of cDNA ends were used to extend the putative cDNA sequences of the mouse and human genes by using rapid amplification of cDNA ends kits according to the manufacturer’s directions (Life Technologies). In addition, IMAGE clones 2063393 [DINB1 expressed sequence tag (EST) AI375146], 1311317 (Dinb1 EST AA920064), and 385429 (Dinb1 EST W62931) were purchased (Research Genetics, Huntsville, AL) and sequenced. PCR products were cloned into vectors pCRII (Invitrogen) or pGEM-T Easy (Promega) by T overhang ligation.
Databases and Protein Sequence Analysis.
The databases used were the nonredundant database of protein sequences and the database of nucleotide sequences of unfinished bacterial genomes at the National Center for Biotechnology Information (National Institutes of Health). The nonredundant database was searched with the gapped blast program and the psi-blast as described (16, 17). The psi-blast program normally was run to convergence, with the e value of 0.01 as the cutoff for including sequences in the profile. Multiple alignments were constructed with the clustalx program (16) and modified manually on the basis of the alignment generated by psi-blast. For phylogenetic tree construction, large inserts and ambiguously aligned regions were removed from the multiple alignment. Phylogenetic trees were constructed by using the neighbor-joining method (18) with 1,000 bootstrap replications as implemented in the phylip package (19).
Chromosome Mapping and Fluorescence in Situ Hybridization.
PCR primers designed to produce a human-specific product from the 5′ end of the DINB1 gene were used to screen the NIGMS human/rodent somatic cell hybrid mapping panel 2. Sequences of the primers were, for forward, 5′-TGGATAGCACAAAGGAGAAGTGTG-3′ and, for reverse, 5′-AATCTGGACCCCTTCGTGGCTTCC-3′. Screening with the PCR primers above yielded a single clone designated pDJ487d14. Fluorescence in situ hybridization was performed as described (20) with biotinylated pDJ487d14 as the probe against normal male donor metaphase chromosomes from cells labeled with BrdUrd for the last 4.5 h of culture (21).
Northern Blot Analysis of DINB1 Expression.
A human multiple-tissue Northern blot II (CLONTECH) containing 2 μg of poly(A)+ RNA/lane was hybridized with a labeled random-primed human DINB1 cDNA probe (nucleotides 659–1,454) according to the manufacturer’s directions.
Reverse Transcription–PCR (RT-PCR) Analysis of DINB1 Expression.
RT-PCR was performed on cDNAs from multiple human tissues by using primers complementary to the 5′ and 3′ ends of the human ORF. The primers used were hDinB-5′ (5′-GTG GAT CCG CCA TGG ATAGCA CAA AGG AGA AGT G-3′) and hDinB-3′ (5′-CAT ACC CTT GAT ATA TTT TTT AAG TAG TCG ACC GCG GAT CCA T-3′). The amount of cDNA used per reaction was as follows: 5 μl of 100 ng/μl HeLa cell library cDNA, 2 μl of 2–10 ng/μl testis cDNA (Origene, Rockville, MD), and 5 μl of 0.2 ng/μl each cDNA from human multiple cDNA panel I (CLONTECH). PCRs were performed by using 2.5 units of Expand High Fidelity DNA polymerase according to the manufacturer’s suggestions (Roche Molecular Biochemicals), and touchdown PCR was performed as described above. Samples (20 μl) were analyzed on a 1% agarose gel in TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3).
Results
cDNA and Protein Sequences of Human and Mouse DinB Homologs.
The human DINB1 sequence of 4,074 nucleotides (GenBank accession no. AF163570) contains an ORF of 2.6 kilobases (kb) that can encode a protein of 870 amino acids with a predicted molecular mass of 99 kDa (Fig. 1). The mouse Dinb1 gene sequence of 4,263 nucleotides (GenBank accession no. AF163571) contains an ORF of 2.55 kb that can encode a protein of 852 amino acids with a predicted molecular mass of 96 kDa (Fig. 1). The context of the translation initiation codon of the human DINB1 ORF (ACCAUGG) is a perfect match to the Kozak consensus sequence (22). That of the mouse Dinb1 ORF (AUCAUGG) is also a good match, especially in the key −4 and +3 positions. The predicted ORFs of the mouse and human genes seem to be complete, because stop codons are present in all three reading frames upstream and downstream of the protein coding regions. Furthermore, the nucleotide sequence identity between the mouse and human genes decreases dramatically immediately outside the putative coding regions, suggesting that these sequences are within the untranslated regions (UTRs).
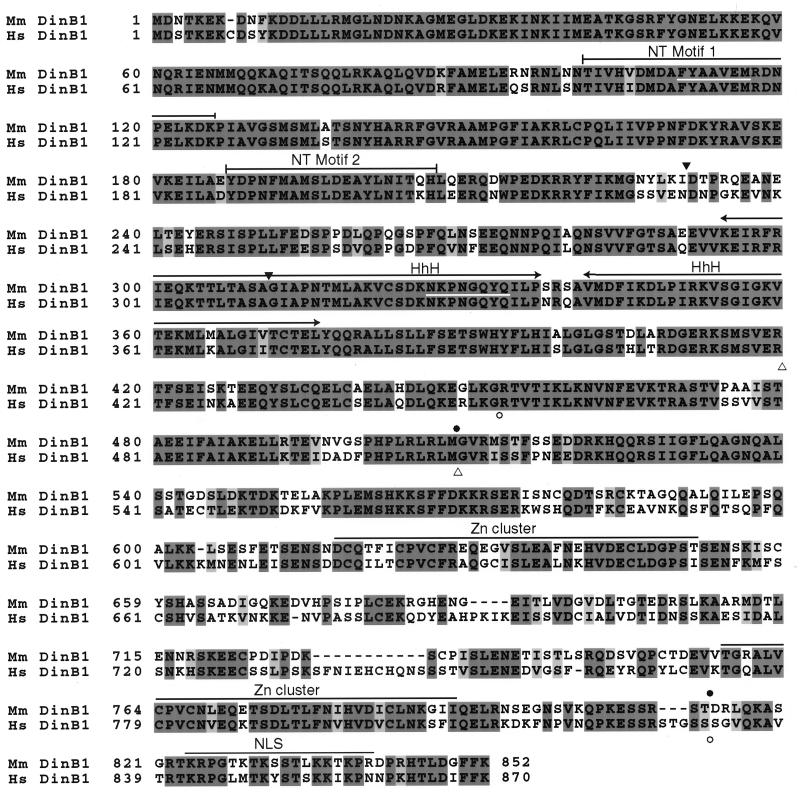
Figure 1.
Amino acid alignment of the mouse (Mus musculus; Mm) and human (Homo sapiens; Hs) DinB proteins. The alignment was generated with clustalw. Identical residues are shaded in dark gray. Similar residues are shaded in light gray. Sites in the mouse DinB1 protein sequence corresponding to the degenerate primers used for cloning are underlined in white (lines 2 and 6 of the paired sequences). Putative functional units are overlined: NT, nucleotidyl transferase; HhH, helix–hairpin–helix; Zn cluster, modified Zn cluster; NLS, bipartite nuclear localization signal. The positions of the predicted deletions in the two isoforms of the mouse [amino acid positions 231–310 (▾) and 509–813 (●)] and human [amino acid positions 420–509 (▵) and 453–831 (○)] DinB1 proteins resulting from alternative splicing of transcripts in testis are indicated.
The sequenced region of the human 3′ UTR contains a putative AAUAAA polyadenylation signal at nucleotide 3,276, which is not always used as a transcriptional termination signal, because additional 3′ UTR sequence is present beyond this point (data not shown). Tissue-specific alternative polyadenylation that uses this signal might account for additional DINB1 transcripts observed in testis by Northern blotting (see Fig. 5). The human DINB1 3′ UTR also contains six copies of the pentanucleotide AUUUA (data not shown); such AU-rich elements, called AREs, have been shown to play a role in destabilization of mRNAs (23). The mouse cDNA is apparently complete, because its size is consistent with the largest mRNA (4.4 kb) detected by Northern analysis (data not shown). The 3′ UTR of the mouse Dinb1 gene contains a consensus AAUAAA polyadenylation sequence at position 4,201 and has 10 copies of the AUUUA destabilization signal (data not shown).
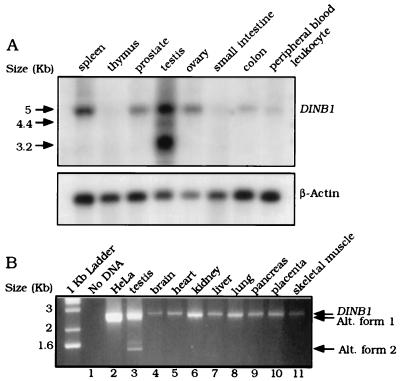
Figure 5.
Expression of the DINB1 gene in human tissues. (A) Northern blot analysis of DINB1 expression in human tissues. A human DINB1 cDNA probe was used to hybridize a multiple-tissue Northern blot (CLONTECH) containing 2 μg of poly(A)+ RNA per lane. A β-actin cDNA probe was used as a control. The size of the full-length DINB1 transcript and the putative alternative transcripts are indicated by arrows on the left. (B) RT-PCR analysis of DINB1 expression in human tissues. RT-PCR was performed on cDNAs from multiple human tissues by using primers complementary to the 5′ and 3′ ends of the ORF. Lanes corresponding to HeLa and testis cDNA are overloaded by comparison with the other lanes. Samples (20 μl) were analyzed on a 1% agarose gel in TBE.
Domain Organization and Phylogenetic Analysis of the UmuC/DinB Superfamily.
The predicted human and mouse DinB1 proteins are substantially hydrophilic (30% acidic/basic residues) and contain bipartite nuclear localization signals at their C termini. The conserved portion of the UmuC/DinB superfamily, including the mammalian DinB homologs, consists of the N-terminal nucleotidyl transferase domain, two tandem HhH domains implicated in DNA binding, and a C-terminal domain of unknown function (Figs. 2 and 3). No sequence similarity between the DinB nucleotidyl transferase domain and other known nucleotidyl transferases/DNA polymerases (or any other enzymes) was detected. (The psi-blast program was run to convergence with a liberal cutoff of e = 0.1 for each member of the superfamily.) However, the multiple alignment of the DinB homologs reveals the presence of two highly conserved motifs that center at an invariant DE doublet (motif 2) and an DXD signature (motif 1) present in most family members (Fig. 2). Both residues of the invariant DE doublet are essential for the DNA polymerase activity of yeast DNA polymerase η (12). Conserved negatively charged residues flanked by hydrophobic residues are a typical feature of many polymerases (24, 25), in which they coordinate divalent cations directly involved in catalysis (26, 27). By inference, a similar role seems likely for the conserved acidic residues of the UmuC/DinB superfamily.
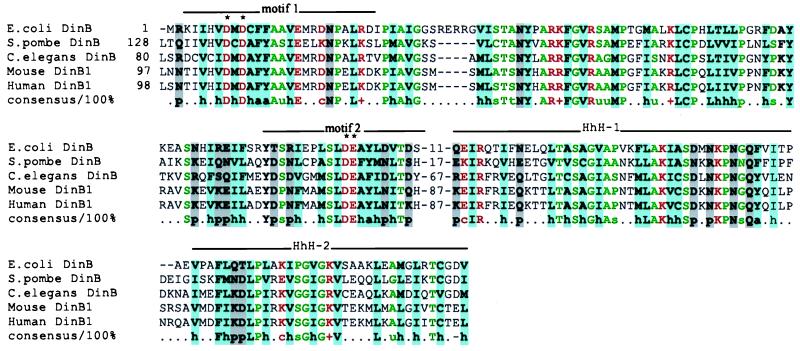
Figure 2.
Conserved motifs within the DinB branch of the UmuC/DinB superfamily. The two conserved motifs containing the putative catalytic residues mentioned in the text (marked by asterisks) and the two HhH modules are overlined. Gene identification numbers are, for E. coli, 2501652; for Schizosaccharomyces pombe, 4038629; and, for C. elegans, 465873. The consensus that includes residues conserved in all aligned sequences is shown beneath the alignment with the following designations: a, aromatic (F, Y, and W); h, hydrophobic (A, C, I, L, V, M, F, Y, and W); p, polar (D, E, N, Q, K, R, H, S, and T); c, charged (D, E, K, and R); +, positively charged (K and R); −, negatively charged (D and E); s, small (G, A, S, P, V, D, and N); and u, tiny (G, A, and S). Conserved residues are color coded according to the consensus.
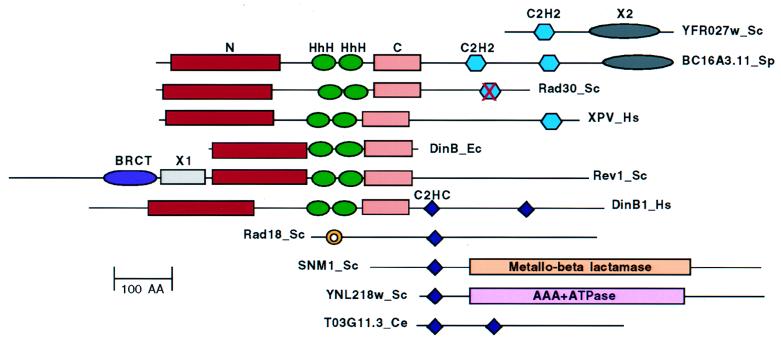
Figure 3.
Domain architecture of the UmuC/DinB superfamily. N, N-terminal nucleotidyl transferase domain; C, conserved C-terminal domain of unknown function; HhH, helix–hairpin–helix; C2HC, Zn cluster; C2H2, Zn finger; X1/X2, uncharacterized domains. The RING domain in Rad18 is designated by a yellow circle. The sequences are grouped by the distinct domain organizations of the respective proteins. Abbreviations: Sc, S. cerevisiae; Sp, S. pombe; Ce, C. elegans; Hs, H. sapiens; Ec, E. coli; 100 AA, 100 amino acids.
The mammalian DinB homologs also contain a duplicated C2HC Zn-cluster domain (Fig. 3). This distinctive version of the Zn finger is present (in combination with other enzymatic and binding domains) in two characterized DNA repair proteins, namely yeast Snm1 (28) and Rad18 (29), the ORC6 subunit of the yeast origin-recognition complex (30), and several uncharacterized proteins (Fig. 3). The apparent orthologs of Snm1 from higher eukaryotes lack the C2HC Zn cluster (L.A. and E.V.K., unpublished observations), underscoring the evolutionary mobility of this domain. Rad18 is a DNA-binding protein with two identifiable distinct domains, a RING finger and the C2HC Zn cluster (29, 31). Because RING domains typically are associated with specific protein–protein interactions (32), it is possible that the Zn cluster is involved in protein–DNA binding. Hence, the mammalian homologs of DinB seem to possess two unrelated DNA-binding domains, the double-HhH domain and the Zn cluster. A C2H2 Zn finger unrelated to the Zn cluster is present in the human XPV protein and its fungal homologs (Fig. 3), although S. cerevisiae Rad30 contains a degenerate version (Fig. 3). This observation underscores the functional association of the UmuC/DinB superfamily nucleotidyl transferases with Zn-binding modules that are likely to provide additional contacts with DNA and shows the plasticity of domain organization of these proteins, a general feature of DNA repair proteins (33).
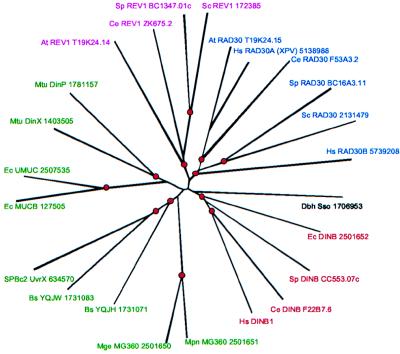
The UmuC/DinB superfamily seems to be represented in all eukaryotes but shows a patchy distribution in bacteria and, thus far, has been identified in only one archaeon, Sulfolobus sofataricus (34). Among bacteria, this family is represented in all Gram-positive bacteria and in some Gram-negative Proteobacteria, but, as yet, the family has not been found in other lineages. Phylogenetic analysis of the UmuC/DinB superfamily reveals several distinct groups that are convincingly supported by the bootstrap test (Fig. 4). These can be separated into four subfamilies exemplified by E. coli UmuC protein, E. coli DinB protein, S. cerevisiae Rev1 protein, and S. cerevisiae Rad30 protein. The Rev1 and Rad30 subfamilies are exclusively eukaryotic, whereas the UmuC subfamily comprises only bacterial proteins (although this finding is not statistically supported as strongly as in the other families). The mouse and human DinB homologs belong to a branch that includes the bacterial DinB protein and its eukaryotic homologs from S. pombe and C. elegans (Fig. 4), suggesting a mitochondrial origin for these eukaryotic genes, with subsequent fusion of the Zn-cluster and the C-terminal globular domains. The presence of N-terminal extensions in the eukaryotic proteins (Fig. 3) that could serve as mitochondrial import peptides is consistent with this interpretation. The phylogenetic position of the DinB homolog from Sulfolobus is uncertain, and in general, it is not possible to propose a definitive evolutionary scenario for this superfamily. Given the presence of the umuC-related mucB genes on plasmids and bacteriophage SPBc2 (35), a major contribution of horizontal gene transfer to the current distribution of the UmuC/DinB superfamily seems likely.
Figure 4.
Unrooted phylogenetic tree of the UmuC/DinB superfamily. The tree was generated from a complete multiple alignment of the UmuC/DinB superfamily of proteins (L.A., unpublished work; available on request). The circles indicate internal nodes with at least 75% bootstrap support. The subfamilies are indicated as follows: UmuC, green; DinB, red; Rad30, blue; and Rev1, pink. The apparent human ortholog of Rev1 was not included, because the sequence is incomplete. Abbreviations are defined in the legend to Fig. 3 and as follows: Dbh Sso, DinB homolog from S. sofataricus; Mpn, Mycoplasma pneumoniae; Mge, Mycoplasma genitalium; Bs, Bacillus subtilis; SPBc2, bacteriophage SPBc2; Mtu, Mycobacterium tuberculosis; and At, Arabidopsis thaliana.
Chromosomal Mapping of the Human DINB1 Gene.
PCR analysis of the NIGMS human/rodent somatic-cell hybrid-mapping panel 2 with primers specific for the human DINB1 cDNA yielded amplification products exclusively in the human control lanes and in the lane for the human chromosome 5/rodent hybrid (data not shown). Fluorescence in situ hybridization with P1 artificial chromosome clone pDJ487d14 containing part of the DINB1 gene yielded a single site of hybridization at band 5q13.1, consistent with the results from the human/rodent hybrid panel screen (data not shown). No cross-hybridization to other homologs was observed. DNA sequencing and PCR analysis showed that the clone contains only the first two exons of the DINB1 gene, plus substantial upstream sequence (data not shown).
Expression of Human DINB1.
The predominant DINB1 transcript observed in human multiple tissue blots is ≈5 kb and is present at low but varying amounts in all tissues examined (Fig. 5A). Expression of the DINB1 gene is highest in testis, with additional abundant transcripts of ≈3.2 and ≈4.4 kb in this tissue. Some of these transcripts might arise because of the alternative use of the polyadenylation signal at position 3,276.
To determine whether alternative splicing occurs within the coding region, the human DINB1 ORF was amplified from cDNA from a number of human tissues. RT-PCR of HeLa cDNA consistently yielded a single product of 2,613 bp identical to the full-length DINB1 ORF reported here (Fig. 5B, lane 2). This 2,613-bp product was also found in a variety of human tissues (Fig. 5B, lanes 4–11). In contrast, RT-PCR of human testis cDNA yielded three products (2,613, 2,344, and 1,484 bp), consistent with possible alternative splicing within the DINB1 coding region (Fig. 5B, lane 3). These cDNA products were cloned, and the putative sites of alternative splicing were mapped (Fig. 1). In the case of human DINB1, both alternate transcripts are expected to result in frameshift mutations.
RT-PCR of mouse testis cDNA with primers to the 5′ and 3′ end of the Dinb1 coding region also results in three products (data not shown). However, the deletions in the mouse Dinb1 alternate transcripts are in frame and are expected to express distinct protein isoforms that retain a nuclear localization signal. Intriguingly, one of the mouse Dinb1 alternative-splice products removes the C-terminal Zn clusters and, hence, may result in a protein with altered DNA-binding activity. At present it is not clear why the putative alternative-splice sites in testis are not conserved between the mouse and human DINB1 genes.
Discussion
The mammalian DinB homologs described here are members of the UmuC/DinB superfamily of replication bypass DNA polymerases (36). The presence of multiple DinB paralogs within single organisms hints that these genes might have overlapping functions or that each is required for the bypass of a particular class of DNA damage. In addition, it seems reasonable to consider that the activity of these DNA polymerases might be error-free for some and error-prone for others, depending on the fidelity of each polymerase and the type of DNA lesion encountered. For example, the Rad30 branch of the UmuC/DinB superfamily seems to represent error-free DNA polymerases, because both the yeast and one of the human homologs (XPV) have the ability to bypass thymine dimers in template DNA accurately (12, 14, 15).
Much of our knowledge about the process of mutagenesis in eukaryotes derives from studies with the yeast S. cerevisiae (1). Unfortunately, this organism seems to lack a true DinB ortholog, although it does have related Rad30 and Rev1 proteins. Hence, it is more difficult to predict the precise catalytic activity of the mammalian DinB proteins described here. However, based on the similarity to E. coli DinB, it seems reasonable to suggest that the mouse and human DinB protein activities participate in an error-prone bypass pathway of DNA replication. The human DINB1 gene might therefore prove to be an oncogene, and its overexpression may lead to tumorigenesis. Chromosome 5q13 has been identified as a frequent site of deletion and translocation in human cancers (37). The latter class of rearrangements is consistent with the hypothesis that DINB1 might be an oncogene.
We have presented evidence that suggests that the expression of the mammalian DinB homologs might be regulated posttranscriptionally, via messenger stability, alternative polyadenylation, and alternative splicing within the coding regions. All of these mechanisms may contribute to regulation of the levels and activity of the DinB homologs. Such regulation may be critical if these proteins increase mutagenesis in cells. Finally, we note that the human DINB1 cDNA is apparently incomplete, because the largest mRNA observed by Northern analysis is ≈5 kb (Fig. 5). We believe that the missing 1 kb of DINB1 cDNA sequence is in the 3′ UTR. Additional ESTs with similarity to the 3′ UTR of the DINB1 cDNA are present in the National Center for Biotechnology Information EST database. However, the 3′ UTR contains a repetitive Alu sequence, and it is not possible to exclude or confirm that these ESTs correspond to the DINB1 gene.
Acknowledgments
We thank Lisiane Meira for providing mouse embryonic fibroblast and mouse testis total RNA, John Feaver and Lurdes Queimado for discussion and critical review of the manuscript, and John McDonald and Roger Woodgate for generously sharing data before publication. V.L.G. is supported by postdoctoral fellowship CA75733 from the National Cancer Institute. This work was supported by National Cancer Institute Grant CA69029 to E.C.F.
Abbreviations
- HhH
helix–hairpin–helix
- UTR
untranslated region
- EST
expressed sequence tag
- RT-PCR
reverse transcription–PCR
- kb
kilobase
Footnotes
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Reuven N B, Tomer G, Livneh Z. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 3.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O’Donnell M, Goodman M F. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohmori H, Hatada E, Qiao Y, Tsuji M, Fukada R. Mutat Res. 1995;347:1–7. doi: 10.1016/0165-7992(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 5.Brotcorne-Lannoye A, Maenhaut-Michel G. Proc Natl Acad Sci USA. 1986;83:3904–3908. doi: 10.1073/pnas.83.11.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S-R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner J, Gruz P, Kim S-R, Yamada M, Matsui K, Fuchs R P P, Nohmi T. Mol Cell. 1999;4:281–287. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 8.Larimer F W, Perry J R, Hardigree A A. J Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 10.McDonald J P, Levine A S, Woodgate R. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roush A A, Suarez M, Friedberg E C, Radman M, Siede W. Mol Gen Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R E, Prakash S, Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 13.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 15.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S F, Koonin E V. Trends Biochem Sci. 1998;11:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 21.Tonk V, Schneider N R, Delgado M R, Mao J-I, Schultz R A. Am J Med Genet. 1996;61:16–20. doi: 10.1002/(SICI)1096-8628(19960102)61:1<16::AID-AJMG3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachs A B. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 24.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braithwaite D K, Ito J. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 27.Saturno J, Lazaro J M, Blanco L, Salas M. J Mol Biol. 1998;283:633–642. doi: 10.1006/jmbi.1998.2121. [DOI] [PubMed] [Google Scholar]
- 28.Richter D, Niegemann E, Brendel M. Mol Gen Genet. 1992;231:194–200. doi: 10.1007/BF00279791. [DOI] [PubMed] [Google Scholar]
- 29.Jones J S, Weber S, Prakash L. Nucleic Acids Res. 1988;25:7119–7131. doi: 10.1093/nar/16.14.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J J, Herskowitz I. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 31.Bailly V, Lauder S, Prakash S, Prakash L. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 32.Borden K L, Freemont P S. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 33.Aravind L, Walker D R, Koonin E V. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulaeva O I, Koonin E V, McDonald J P, Randall S K, Rabinovich N, Connaughton J F, Levine A S, Woodgate R. Mutat Res. 1996;357:245–253. doi: 10.1016/0027-5107(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 35.Woodgate R, Sedgwick S G. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 36.Friedberg E C, Gerlach V L. Cell. 1999;98:413–416. doi: 10.1016/s0092-8674(00)81970-4. [DOI] [PubMed] [Google Scholar]
- 37.Mitelman F, Mertens F, Johannson B. Nat Genet. 1997;15:417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]