Abstract
The AtTOM1 gene of Arabidopsis thaliana had been shown to be essential for the efficient multiplication of Tobacco mosaic virus (TMV) in A. thaliana. In this study, we cloned an AtTOM1-like gene from Nicotiana benthamiana named as NbTOM1. Sequence alignment showed that NbTOM1 is closely related to AtTOM1 homologues of N. tabacum and Lycopersicon esculentum with 97.2% and 92.6% nucleotide sequence identities, respectively. Silencing of NbTOM1 by a modified viral satellite DNA-based vector resulted in complete inhibition of the multiplication of TMV in N. benthamiana. The result suggests that inhibition of NbTOM1 via RNA silencing is a potentially useful method for generating TMV-resistant plants.
Keywords: Tobacco mosaic virus, NbTOM1, Virus induced gene silencing, Nicotiana benthamiana
INTRODUCTION
Viral diseases have caused significant yield and quality losses in crop production. Over the past years, transgenic approaches have successfully generated virus-resistant plants (Goldbach et al., 2003). An effective method for obtaining resistant transgenic plants is to induce RNA silencing by expressing virus-derived dsRNA in plants (Zhang et al., 2005).
A variety of host factors were identified to be involved in the intracellular multiplication of viruses (Lai, 1998; Lee et al., 2001; Noueiry et al., 2000). Previous studies showed that AtTOM1 and AtTOM3 of Arabidopsis thaliana, which encode a putative seven-pass transmembrane protein, are essential for the efficient multiplication of a crucifer-infecting tobamovirus (TMV-Cg) (Hagiwara et al., 2003; Yamanaka et al., 2000; 2002). Two AtTOM1 homologues, NtTOM1 and NtTOM3, were identified in Nicotiana tabacum. Simultaneous RNA interference against NtTOM1 and NtTOM3 in N. tabacum successfully inhibited the multiplication of Tomato mosaic virus and other tobamoviruses (Asano et al. 2005). AtTOM1 and AtTOM3 homologues are present in various plant species and their inhibition via RNA interference should constitute a useful method for generating tobamovirus-resistant plants (Asano et al., 2005). In this study, we identified an AtTOM1-like gene in N. benthamiana, named as NbTOM1 and reported that silencing of NbTOM1 by a modified viral satellite DNA-based vector results in complete inhibition of Tobacco mosaic virus (TMV) multiplication in N. benthamiana.
MATERIALS AND METHODS
Isolation of TOM1 homologue and construction of gene silencing vector
To identify TOM1 homologue in N. benthamiana, total RNA from N. benthamiana leaves was prepared using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and applied as a template for first-strand cDNA synthesis using oligo d(T)18 and superscript reverse transcriptase (TaKaRa, Shiga, Japan). The TOM1 homologue fragment was PCR-amplified with the degenerated primers TOM1F (5′ CTTTGTYGTVAAYGGAGTTC 3′) and TOM1R (5′ GTAYTGWGCWGAYACTC 3′) (V=A, C, G; W=A, T; Y=C, T) using the synthesized first-strand cDNA as template. The resulting PCR product was cloned into pGEM-Teasy vector (Promega, Madison, WI, USA) and then sequenced. Based on the fragment sequence, primers TOM1F1 (5′ TCTAGACTATTGTCTTTGGATTTCACA 3′, restriction site for BamHI is underlined) and TOM1R1 (5′ GGATCCAGGCAATTTTCGCAGGATG 3′, restriction site for XbaI is underlined) were designed. The resulting PCR product was digested with BamHI-XbaI and inserted into the BamHI-XbaI-cut DNAβ vector of Tomato yellow leaf curl China virus (TYLCCNV) (DNAmβ) (Tao and Zhou, 2004) to produce construct DNAmβ:NbTOM1. The infectious clone of DNAmβ and TYLCCNV was constructed previously by the authors’ lab (Cui et al., 2004; Tao and Zhou, 2004).
Agroinoculation of plants
DNAmβ:NbTOM1 were introduced into Agrobacterium tumefaciens strain PGV3850 by electropolation. A. tumefaciens cultures containing infectious clone of TYLCCNV and DNAmβ:NbTOM1 were injected into stems of N. benthamiana seedlings at six-leaf stage as previously described (Zhou et al., 2003). Plants co-inoculated with TYLCCNV and DNAmβ empty vector or water were used as control. Inoculated plants were grown in an insect-free cabinet at a constant temperature of 25 °C with supplementary lighting for 16 h per day.
Virus inoculation
TMV:GFP (green fluorescent protein) carrying the mGFP4 gene was kindly provided by the Sainsbury Laboratory of John Innes Centre. The virus was agro-infiltrated into the sixth true leaf.
GFP image
GFP was detected visually in intact plants with the aid of a hand-held 100 W long-wave ultraviolet lamp (UV products, Upland, CA, USA). Plants were photographed with a Nikon 5 000 digital camera and the images were processed using Adobe Photoshop software.
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was performed as described by Tao and Zhou (2004). cDNA derived from the equivalent of 1 µg of total RNA was used as a template. Primers (TOM1F, TOM1R) that anneal outside the region targeted for silencing were used to ensure that only the endogenous gene transcript was assayed. The elongation factor 1-alpha (EF-1-α) served as an internal control for RNA quantity in RT-PCR. The intensities of PCR-generated fragments were analyzed and quantified using Gel-Pro Analyzer 3.1 (Media Cybernetics, Atlanta, GA, USA).
RNA gel blot analysis
Total RNA isolation using systemic leaves and Northern blot analysis were conducted as described previously (Cui et al., 2004). Membranes were hybridized with [α-32P]dCTP-labelled probe specific for the TMV movement protein gene. Hybridization signals were detected by phosphorimaging using a Typhoon 9200 imager (Amersham Pharmacia, Piscataway, NJ, USA).
RESULTS AND DISCUSSION
AtTOM1 homologues were found in Lycopersicon esculentum (AB193043) Oryza sativa (AK066373) and N. tabacum (AB193039). Based on the alignment of nucleotide sequences of these homologues, we designed degenerate primers TOM1F and TOM1R. A 0.6 kb cDNA fragment was PCR-amplified with these primers from N. benthamiana. Sequence analysis and comparison revealed that the fragment (AM261863, named as NbTOM1 gene) was closely related to AtTOM1 homologues of N. tabacum and L. esculentum with 97.2% and 92.6% nucleotide sequence identities, respectively (Table 1).
Table 1.
Nucleotide (bottom left) and amino acid (top right) sequence identities of NbTOM1 with AtTOM1 and TOM1 homologues of Lycopersicon esculentum (LeTOM1), Oryza sativa (OsTPM1) and Nicotiana tabacum (NtTOM1)
| NbTOM1 | NtTOM1 | LeTOM1 | AtTOM1 | OsTPM1 | |
| NbTOM1 | − | 97.4 | 94.3 | 81.3 | 74.1 |
| NtTOM1 | 97.2 | − | 93.8 | 77.4 | 71.9 |
| LeTOM1 | 92.6 | 80.3 | − | 76.0 | 71.2 |
| AtTOM1 | 69.5 | 59.0 | 52.5 | − | 71.8 |
| OsTPM1 | 63.9 | 80.3 | 47.2 | 49.9 | − |
To investigate whether NbTOM1 supports TMV multiplication, we tried to inhibit the expression of NbTOM1 by virus induced gene silencing (VIGS). For this purpose, the NbTOM1 fragment were introduced into the TYLCCNV DNAβ-based gene silencing vector (Tao and Zhou, 2004) to generate DNAmβ:NbTOM1. N. benthamiana plants were then co-agroinoculated with TYLCCNV and DNAmβ: NbTOM1, TYLCCNV and DNAmβ empty vector or water. Approximately 15 d after co-agroinoculation, plants were challenge-inoculated with the TMV:GFP. Within 3 weeks after challenge-inoculation, all the six tested plants that were co-agroinoculated with TYLCCNV and DNAmβ:NbTOM1 yielded a red signal under UV (Fig.1a), due to non-proliferation of the TMV:GFP clone. In contrast, the control plants yielded a green signal due to the GFP expression accompanying TMV multiplication (Figs.1b and 1c). When NbTOM1-silenced N. benthamiana plants were challenge-inoculated with Cucumber mosaic virus, the inoculated plant produced severe viral symptoms similar to those in non-silenced control plants (data not shown). These results suggest that the multiplication of TMV is inhibited in the NbTOM1 silenced plants.
Fig. 1.



Effects of the knockdown of NbTOM1 expression on TMV accumulation in N. benthamiana. Plants were pre-inoculated with TYLCCNV and DNAmβ: NbTOM1 (a), TYLCCNV and DNAmβ empty vector (b) and water (c), respectively, and then challenge inoculated with the TMV:GFP clone 15 d after pre-inoculation. The NbTOM1 silenced plant appear red due to non-proliferation of TMV:GFP (a). The NbTOM1 non-silenced control plant appear green due to GFP expression resulting from TMV:GFP accumulation (b, c). Photographs were taken under UV light 3 weeks after challenge inoculation
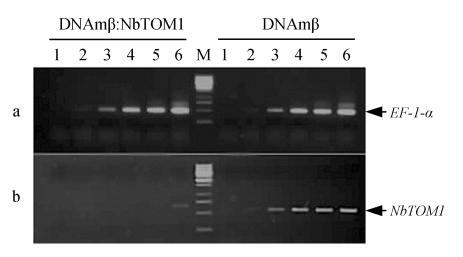
To assay the mRNA levels from the endogenous NbTOM1 gene, we performed semi-quantitative RT-PCR. The results obtained revealed that the level of NbTOM1 mRNA was reduced by more than 80% in the systemic NbTOM1 silenced N. benthamiana plants compared with the control plants. In contrast, the level of mRNA of EF-1-α, which served as an internal control for RNA quantity in RT-PCR amplification, was similar in NbTOM1-silenced and the control plants (Fig.2). These results suggest that NbTOM1 mRNA is a target of VIGS.
Fig. 2.
Semi-quantitative RT-PCR analysis showing the effect of VIGS with DNAmβ:NbTOM1 in N. benthamiana. Lanes 1~6 correspond to products from PCR cycle Nos. 15, 18, 21, 24, 27 and 30. Lane M represents size markers. (a) PCR products for EF-1-α derived from N. benthamiana co-agroinfected with TYLCCNV and DNAmβ:NbTOM1 (left) or with TYLCCNV and DNAmβ empty vector (right). Gene EF-1-α served as an internal control for RNA quantity in RT-PCR; (b) PCR products of NbTOM1 from N. benthamiana co-agroinoculated with TYLCCNV and DNAmβ: NbTOM1 (left) or with TYLCCNV and DNAmβ empty vector (right)
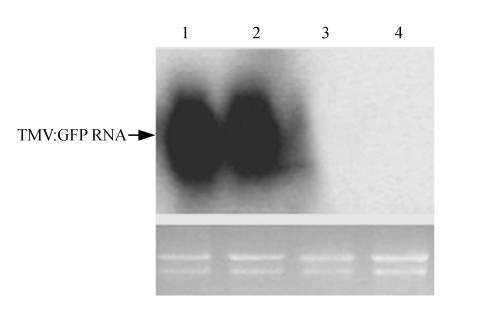
To further monitor the level of TMV multiplication, total RNA was isolated from leaves and analyzed by Northern blot hybridization using a probe specific for TMV. Hybridization results revealed a complete inhibition of TMV multiplication in NbTOM1 silenced plants, correlating well with the results from the corresponding GFP expression assays (Fig.3).
Fig. 3.
Northern blot analysis of TMV accumulation in NbTOM1 silenced and non-silenced N. benthamiana plants at 18 d post-inoculation with TMV:GFP
Lanes 1 and 2 show viral RNA from NbTOM1-non-silenced plants; Lanes 3 and 4 show viral RNA from individual NbTOM1-silenced plants. Lower panels show RNAs stained with ethidium bromide
The discovery of host factors, including proteins, membranes, and nucleic acids that are implicated in viral infection cycles is providing fundamental clues about the molecular bases of viral susceptibility. As a host factor, AtTOM1, which is associated with the TMV-Cg replication complex, supports efficient TMV-Cg multiplication in A. thaliana (Yamanaka et al., 2000). In this report, we characterized an AtTOM1-like gene from N. benthamiana (NbTOM1), and demonstrated that NbTOM1 plays an essential role in TMV multiplication. Our results are consistent with previous reports.
We have demonstrated that the VIGS-induced knockdown of NbTOM1 confers strong TMV resistance in N. benthamiana. As AtTOM1 homologues have been reported in several kinds of plants (Asano et al., 2005), similar strategies might be applicable to a wide range of economically important plants. Compared with conventional transgenic approaches, VIGS offers advantages for generating virus-resistant plants because it does not rely on stable plant transformants.
Footnotes
Project supported by the Cultivation Fund of the Key Scientific and Technical Innovation Project, Ministry of Education of China (No. 705025) and the National Natural Science Foundation of China (No. 30530520)
References
- 1.Asano M, Satoh R, Mochizuki A, Tsuda S, Yamanaka T, Nishiguchi M, Hirai K, Meshi T, Naito S, Ishikawa M. Tobamovirus-resistant tobacco generated by RNA interference directed against host genes. FEBS Lett. 2005;579(20):4479–4484. doi: 10.1016/j.febslet.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 2.Cui XF, Tao XR, Xie Y, Fauquet CM, Zhou XP. A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J Virol. 2004;78(24):13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldbach R, Bucher E, Prins M. Resistance mechanisms to plant viruses: an overview. Virus Res. 2003;92(2):207–212. doi: 10.1016/S0168-1702(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 4.Hagiwara Y, Komoda K, Yamanaka T, Tamai A, Meshi T, Funada R, Tsuchiya T, Naito S, Ishikawa M. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 2003;22(2):344–353. doi: 10.1093/emboj/cdg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai MMC. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244(1):1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 6.Lee WM, Ishikawa M, Ahlquist P. Mutation of host Delta 9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J Virol. 2001;75(5):2097–2106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noueiry AO, Chen JB, Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc Natl Acad Sci USA. 2000;97(24):12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao XR, Zhou XP. A modified viral satellite DNA that suppresses gene expression in plant. Plant J. 2004;38(5):850–860. doi: 10.1111/j.1365-313X.2004.02087.x. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka T, Ohta T, Takahashi M, et al. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA. 2000;97(18):10107–10112. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamanaka T, Imai T, Satoh R, Kawashima A, Takahashi M, Tomita K, Kubota K, Meshi T, Naito S, Ishikawa M. Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J Virol. 2002;76(5):2491–2497. doi: 10.1128/jvi.76.5.2491-2497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, Niu YB, Zhou XP. Transgenic tobacco plants expressing dsRNA can prevent infection by Tobacco mosaic virus . Chin J Agric Biotechnol. 2005;2(3):201–205. doi: 10.1079/CJB200578. [DOI] [Google Scholar]
- 12.Zhou XP, Xie Y, Tao XR, Zhang ZK, Li ZH, Fauquet CM. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J Gen Virol. 2003;84(1):237–247. doi: 10.1099/vir.0.18608-0. [DOI] [PubMed] [Google Scholar]