Abstract
Background
Insecticide resistance in malaria vector mosquitoes presents a serious problem for those involved in control of this disease. South Africa experienced a severe malaria epidemic during 1999/2000 due to pyrethroid resistance in the major vector Anopheles funestus. Subsequent monitoring and surveillance of mosquito populations were conducted as part of the malaria vector control programme.
Methods
A sample of 269 Anopheles funestus s.l. was collected in Mamfene, northern KwaZulu-Natal, using exit window traps in pyrethroid sprayed houses between May and June 2005. Mosquitoes were identified to species level, assayed for insecticide susceptibility, analysed for Plasmodium falciparum infectivity and blood meal source.
Results
Of the 220 mosquitoes identified using the rDNA PCR method, two (0.9%) were An. funestus s.s. and 218 (99.1%) Anopheles parensis. Standard WHO insecticide susceptibility tests were performed on F1 progeny from wild caught An. parensis females and a significant survival 24 h post exposure was detected in 40% of families exposed to 0.05% deltamethrin. Biochemical analysis of F1 An. parensis showed no elevation in levels/activity of the detoxifying enzyme systems when compared with an insecticide susceptible An. funestus laboratory strain. Among the 149 female An. parensis tested for P. falciparum circumsporozoite infections, 13.4% were positive. All ELISA positive specimens (n = 20) were re-examined for P. falciparum infections using a PCR assay and none were found to be positive. Direct ELISA analysis of 169 blood meal positive specimens showed > 75% of blood meals were taken from animals. All blood fed, false positive mosquito samples for the detection of sporozoites of P. falciparum were zoophilic.
Conclusion
The combination of pyrethroid resistance and P. falciparum false-positivity in An. parensis poses a problem for vector control. If accurate species identification had not been carried out, scarce resources would have been wasted in the unnecessary changing of control strategies to combat a non-vector species.
Background
Historical studies have shown that Anopheles funestus and Anopheles arabiensis are the principal malaria vectors in southern Africa [1,2]. The Anopheles funestus group includes nine African species: An. funestus, Anopheles rivulorum, Anopheles vaneedeni, Anopheles leesoni, Anopheles confusus, Anopheles fuscivenosus, Anopheles brucei, Anopheles parensis and Anopheles aruni. Recently, "An. rivulorum-like" was added to this group based on molecular sequencing data [3]. These species show morphological overlap, although some species can be identified on egg and larval characteristics [1,2]. Of these, An. funestus is the only member of the group that is recognized as an important vector of malaria in Africa, with An. rivulorum only a minor vector at a localized site in Tanzania [4]. Anopheles vaneedeni was experimentally infected in the laboratory with Plasmodium falciparum but has not been implicated in malaria transmission in nature [5].
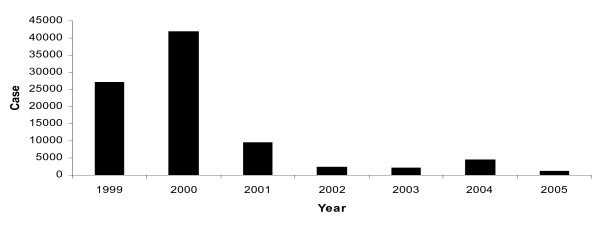
Mamfene village (Figure 1), in northern KwaZulu-Natal Province, South Africa, is situated in an area that was worst affected by the malaria epidemic of 1999/2000 [6]. The seven-fold increase of malaria cases compared with 1994 was mainly linked to the recurrence of An. funestus which was shown to have developed resistance to pyrethroid insecticides [6,7]. A policy decision to reintroduce DDT for malaria control in traditional style houses whilst continuing to use pyrethroids in western style houses resulted in a significant decrease in the number of reported malaria cases in the region during the period 2001 to date (Figure 2). Continued monitoring of vectors within this region has subsequently demonstrated DDT resistance in An. arabiensis (another major malaria vector) and Anopheles quadriannulatus (a non vector) during 2002 [8]. The latter two species are both members of the Anopheles gambiae complex.
Figure 1.
Map of South Africa showing the collection site, Mamfene, within KwaZulu-Natal.
Figure 2.
Reported malaria cases in KZN (1999–2005). Data were obtained from the Jozini Department of Health (unpublished).
Monitoring of malaria vector species continued during 2005 as support for the malaria control programme in northern KwaZulu-Natal. Field collections produced large numbers of the An. funestus group collected in pyrethroid sprayed houses in Mamfene.
The aim of this study, therefore, was to determine the species composition of the An. funestus group in Mamfene, the P. falciparum infection rates in these species, their feeding behavior and their susceptibility to insecticides.
Materials and methods
Study site
Mosquitoes were collected in Mamfene, northern KwaZulu-Natal, South Africa (27°23' S, 32°12' E). Mamfene is an undeveloped rural and rice growing area near the Balamhlanga river that forms an extensive marsh during the rainy season (October to February). Marshy conditions may persist during the dry season when the rice paddies upstream are drained. In addition to growing rice, inhabitants of this area also grow vegetables and cotton, and keep livestock for subsistence. Two types of houses are found in the area: traditional houses, which are mud structures, and western-style houses built using cement and bricks. During 2005, vector control was conducted by spraying a combination of DDT in traditional houses and pyrethroid (deltamethrin) in the westernized houses.
Mosquito collections
Window exit traps were used to collect adult mosquitoes indoors between May and June 2005. Collections were conducted by the KwaZulu-Natal Department of Health, Malaria Control Programme teams. Live mosquitoes were identified using morphological keys [1]. Mosquitoes identified as An. funestus group were transported to the National Institute for Communicable Diseases (NICD) in Johannesburg. Live, blood-fed female anophelines were individually isolated for oviposition and kept under standard insectary conditions (25°C, 75–80% relative humidity and 12 hrs light: dark with 45 min dusk/dawn transition). Larvae were reared through to adults. The F1 progeny (1–4 days old) were subjected to standard WHO insecticide susceptibility tests [9]. Unexposed males and females (1–4 day old) from large families were frozen at -70°C for biochemical analysis. Dead wild-caught females were preserved on silica gel for species identification and sporozoite detection by ELISA.
Species-specific identification
Wild-caught mosquitoes were individually identified using PCR [10]. Two legs per mosquito were used for the assay.
Sporozoite detection by indirect ELISA
The heads and thoraces of wild female mosquitoes were tested for the presence of P. falciparum circumsporozoite protein (CSP) using monoclonal antibodies 2A10 [11]. Seven negative controls and one positive control were included on each plate. The negative controls consisted of triturated, unfed An. funestus s.s. colonized in the Botha De Meillon Insectary at the NICD. The positive control consisted of a synthetic peptide standardized against P. falciparum. Results were analysed using a microtitre plate reader at a wavelength of 405 nm. The absorbance cut off value for positive specimens was calculated as twice the mean value of the four negative controls. Specimens that tested positive for P. falciparum were re-examined using PCR [12] in order to confirm the results.
Polymerase chain reaction (PCR) in order to confirm indirect ELISA positive specimens
Phenol-chlorophorm was used to extract DNA from the positive ELISA homogenate [13]. Two Plasmodium- specific primers based on the sequence of the small subunit ribosomal RNA (ssrRNA) namely rPLU (5 and 6) and rFAL (1 and 2) were used [12]. The rPLU and rFAL primers were separately used for the first and second round of PCR. Briefly, 1 μl was used for PCR [12] with the following conditions: 95°C for 5 min followed by 25 cycles at 94%°C for 1 min, 58°C for 2 min, 72°C for 2 min and a final auto-extension at 72°C for 5 min. One microlitre of the first round PCR was used as template for the second PCR.
Mosquito blood sources (direct ELISA)
Half-gravid and fully fed mosquitoes that were not used for egg-laying were preserved on silica gel. These were screened for blood meal source using direct ELISA method [14]. All specimens were screened for human, bovine, chicken, goat and sheep blood. Absorbance at 405 nm was recorded with an ELISA plate reader 30 min after the addition of the substrate. Samples were considered positive if absorbance values exceeded the mean plus three times the standard deviation of four negative controls (unfed colony mosquitoes).
Insecticide susceptibility tests
Standard insecticide treated papers supplied by WHO [9] were used to test for susceptibility/resistance to 0.1% bendiocarb, 4% DDT, 0.05% deltamethrin and 0.75% permethrin. Anopheles quadriannulatus (SKUQUA), known to be susceptible to all insecticides, was used as a control to ensure the reliability of each batch of impregnated papers. Negative controls consisted of untreated papers. Knockdown was recorded after 1 h and a 10% sucrose solution was made available to survivors. Final mortality was scored 24 h post exposure.
Biochemical analysis
Assays designed to quantify average levels of monooxygenase, non-specific esterase and glutathione-S- transferase (GST) activity as well as to detect the presence, in individual mosquitoes, of an altered acetylcholinestrase associated with carbamate/organophosphate resistance were performed [15]. Propoxur was used as a reference carbamate insecticide for the acetylcholinesterase assay. Mixed samples of male and female F1 progeny from familial material initially stored at -70°C were assayed concurrently with male and female samples drawn from an An. funestus laboratory strain (designated FANG) which is fully susceptible to insecticides. Optical densities were directly compared between the An. parensis and FANG samples by gender following adjustment for total protein content.
Results
Mosquito collections
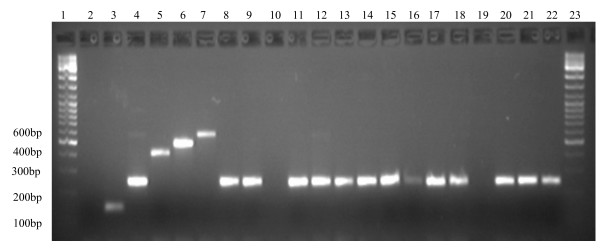
A total of 269 mosquitoes morphologically identified as members of the An. funestus group were collected from window exit traps. Of these, 220 were identified to species level by PCR. Two species were identified: An. parensis 99.1% (n = 218) and An. funestus s.s 0.9% (n = 2). The remaining specimens (n = 49) could not be identified despite three attempts, although the positive controls amplified successfully (Figure 3).
Figure 3.
Species identification of members of An. funestus group from Mamfene. Lanes 1&23: 100 bp (HyperLadder IV from Bioline); Lane2: negative control; Lanes 3–7: positive controls with 3: An leesoni 146 bp, 4: An. parensis 252 bp, 5: An. rivulorum 411 bp, 6: An. funestus 505 bp and 7: An. vaneedeni 587 bp. Lanes 8–9, 11–18 and 20–22 An. parensis from Mamfene. Lanes 10 and 19 did not amplify.
Sporozoite detection
Of 149 female An. parensis assayed using the ELISA method, 13.4 % (20/149) tested positive for P. falciparum circomsporozoites. Those females identified as An. funestus (n = 2) were negative. This assay was repeated three times and the results concurred. Owing to this abnormally high infection rate, all positive specimens were retested using a PCR-based method [12] in an effort to confirm these results. None of the specimens re-examined showed P. falciparum amplicons even though the positive control amplified successfully with each reaction.
Blood meal identification
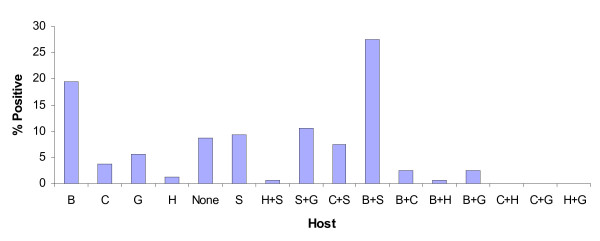
A total of 169 mosquitoes identified by PCR (167 An. parensis & 2 An. funestus) had taken blood meals prior to collection. Both the An. funestus specimens had taken bovine blood. Results for the An. parensis sample are summarized in Figure 4 which shows a broad spectrum of blood meal sources. These include: bovine 19.37%, sheep 9.37% goat 5.62%, chicken 3.75% and human 1.25%. Multiple blood meal sources were detected in An. parensis: 27.5% were positive for bovine and sheep blood and only 0.62% was positive for human and goat blood. Identification of each source of blood was done in duplicate. The remaining 8.75% of blood fed An. parensis could not be assigned a blood meal source.
Figure 4.
Percentages of different and mixed blood meals for Anopheles parensis from Mamfene. B = bovine, C = chicken, H = human, G = goat and S = sheep.
Of the 13 % of females that tested positive for the presence of P. falciparum sporozoites, 65 % (13/20) were blood fed. Identification of blood meal source of those females by direct ELISA showed that 53.85 % were positive for bovine blood, 38.46 % for sheep blood and 7.69 % had a mixed blood meal source.
WHO susceptibility tests
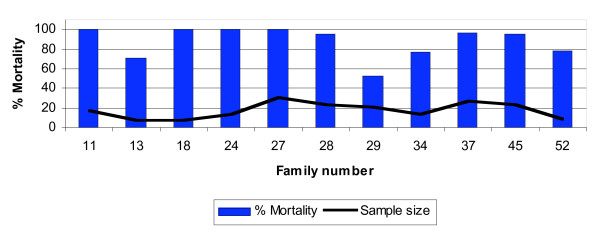
Table 1 shows the percentage mortality recorded 24 h post exposure of 1–4 day old F1 progeny of wild caught An. parensis females. They were assayed against four insecticides: two pyrethroids (deltamethrin and permethrin), one carbamate (bendiocarb) and one organochlorine (DDT). These samples were susceptible to permethrin, bendiocarb and DDT but showed signs of resistance to deltamethrin. Significant survival 24-hr post exposure (> 20%), was detected in 4 of 11 families exposed to 0.05% deltamethrin (Fig. 5) with one family (#29) giving 47.6% survival. All positive controls were 100% susceptible to the insecticides and all negative controls on untreated papers survived.
Table 1.
Results of insecticide susceptibility tests on F1 progeny from families of wild-caught Anopheles parensis 24 hrs post-exposure to three classes of insecticides.
| Insecticide | No. families | TotalNo. | Alive | Dead | % Mortality |
| 0.1% Bendiocarb | 10 | 205 | 4 | 201 | 98.05 |
| 4% DDT | 4 | 81 | 0 | 81 | 100 |
| 0.75% Permethrin | 6 | 118 | 2 | 116 | 98.31 |
| 0.05% Deltamethrin | 11 | 204 | 30 | 174 | 85.29 |
| Control | 100 | 0 | 100 | 100 |
Control = 25 susceptible laboratory Anopheles quadriannulatus colony mosquitoes exposed to each insecticide.
Figure 5.
Percentage mortality 24 h post exposure to0.05% deltamethrin of F1 progeny from An. parensis families.
Biochemical analysis
Direct comparison of optical density values between the An. parensis and laboratory An. funestus male and female samples are summarized in Table 2. Two-sample t tests assuming equal variances revealed no significant differences in detoxifying enzyme levels between strains by gender with the exception of the α esterase assay in which the An. funestus female sample showed a significantly higher mean optical density (OD) value compared to the corresponding An. parensis sample. Male (n = 24) and female (n = 24) samples from both strains showed > 55% inhibition of acetylcholinesterase activity when challenged with propoxur with the exception of one An. parensis female that showed 33.3% inhibition.
Table 2.
Mean optical density values for glutathione S-transferase (GST), monooxygenase (Oxy) and esterase (Est) enzymes using α and β napthyl acetate as substrates.
| n | GST | Oxy | αEst | βEst | ||
| Females | An. parensis | 24 | 0.024 (0.003) | 0.335 (0.022) | 0.134 (0.018) | 0.115 (0.016) |
| An. funestus (FANG) | 24 | 0.022 (0.002) | 0.324 (0.016) | 0.201 (0.026) | 0.16 (0.021) | |
| p | 0.73 | 0.69 | 0.04 | 0.1 | ||
| Males | An. parensis | 24 | 0.024 (0.003) | 0.245 (0.007) | 0.173 (0.023) | 0.178 (0.017) |
| An. funestus (FANG) | 24 | 0.024 (0.002) | 0.246 (0.01) | 0.207 (0.027) | 0.14 (0.018) | |
| p | 0.86 | 0.94 | 0.35 | 0.16 |
"n" = sample size; Figures in parentheses are the standard errors for each sample; p indicates significance in difference between An. parensis and An. funestus samples by enzyme system by gender following 2-sample t tests.
Discussion
Little is known of the vectorial capacity of An. parensis and it is generally regarded as zoophilic with no medical importance [1,2]. Anopheles parensis is found in eastern Africa from Kenya and Tanzania in the north to KwaZulu-Natal Province in South Africa [1,2]. In KwaZulu-Natal, An. parensis has previously been found resting indoors in formerly insecticide sprayed localities (insecticide not mentioned, but DDT traditionally used for house spraying in South Africa) [2]. More recent work in the same area [7,10] also found An. parensis resting inside insecticide sprayed houses. In both these studies failure to identify a proportion of An. funestus group collections could be the result of DNA degradation due to poor preservation or the current lack of species specific primers for all members of this group. The PCR assay [10] only identifies the five most common members and cannot exclude the possibility that other members of the group might be amongst the specimens that failed to be identified. It is suggested that unidentified An. funestus group specimens from Kenya (10%) could have been An. confusus [16]. Anopheles confusus adults are morphologically similar to An. funestus s.s. [2] but cannot be identified to species level using current molecular methods. This rare species has been recorded in Zimbabwe but is generally confined to the plateau areas of eastern Africa from Kenya to South Africa [2].
Recent studies in the Mwea area of central Kenya showed high numbers of An. parensis resting inside houses [16]. Of these only 1.4% had fed on human blood and none were sporozoite positive [16]. The present study showed that 13.5% of the specimens tested were positive for P. falciparum circumsporozoite protein using monoclonal antibodies [11] but only 1.25% had fed on humans. Re-analysis using PCR [12] revealed no positive specimens in this sample. False positive results have been recorded previously [17-19]. A cross-reactive factor (s) in plasma from swine and bovine blood has been postulated to interfere with the ELISA technique resulting in false positives in An. dirus when testing for P. falciparum and P. vivax sporozoites [19]. False positive results have also been reported in An. gambiae tested for P. falciparum antigens [18] and in An. marshallii s.l. [17].
None of the 65 % (13/20) false positive An. parensis specimens that were blood fed had fed on humans. The remaining 35% of positives (7/20) did not show any indication of a blood meal, but still gave false positive results. These false positives could be due to traces of animal blood accumulating in the haemolymph [19]. Why the South African An. parensis should give high false positive results but the Kenyan samples did not [9] needs further investigation.
F1 Progeny from the An. parensis families were susceptible to DDT, permethrin and bendiocarb but showed significant resistance to 0.05% deltamethrin according to WHO criteria [9] in some families, with an overall 24-hr mortality of 85.3%. However, preliminary biochemical analysis showed no detoxifying enzyme systems that could be associated with deltamethrin resistance.
Conclusion
The combination of insecticide resistance and false positive P. falciparum infections in An. parensis underlines yet again the importance of accurate species identification in a vector control monitoring and surveillance programme. Without accurate species identification, scarce resources would have been wasted on changing vector control intervention strategies to target a species that is not feeding on humans and is not transmitting malaria parasites. Eighty years after Watson coined the term "species sanitation" to describe the process of targeting only disease transmitting vectors [20], the concept remains as valid today as it did in those early days of malaria control.
Authors' contributions
JC carried out the rearing of mosquitoes, all the bioassays including ELISAs and molecular studies, wrote the first and subsequent drafts of the manuscript.
K took charge of the field work and morphological identification of anophelines.
RH assisted in interpretation of bioassay results and field work data.
SV performed the biochemistry assays.
BD interpreted the results of the biochemistry assays and contributed to the subsequent writing of the manuscript.
LL and M conceived the project, oversaw its implementation and contributed to the subsequent writing of the manuscript.
All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Prof R. A. Wirtz (CDC Entomology Branch, Atlanta, GA) for supplying P. falciparum-positive controls and monoclonal antibody P. falciparum 2A10 for indirect ELISA used in this study. This investigation received financial support under the Multilateral Initiative on Malaria (MIM) project A 40036 through the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR) to LLK. This work was also partially funded by the South African Medical Research Council and the National Health Laboratory Service Research Trust to LLK.
Contributor Information
Joel C Mouatcho, Email: joelm@nicd.ac.za.
Keith Hargreaves, Email: maureenc@nicd.ac.za.
Lizette L Koekemoer, Email: lizettek@nicd.ac.za.
Basil D Brooke, Email: basilb@nicd.ac.za.
Shüne V Oliver, Email: shüneo@nicd.ac.za.
Richard H Hunt, Email: maureenc@nicd.ac.za.
Maureen Coetzee, Email: maureenc@nicd.ac.za.
References
- Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara. Vol. 55. South African Institute for Medical Research; 1987. pp. 33–81. [Google Scholar]
- Gillies MT, De Meillon B. The Anophelinae of Africa south of the Sahara. Vol. 54. South African Institute for Medical Research; 1968. pp. 131–132. [Google Scholar]
- Cohuet A, Simard F, Toto JC, Kengne P, Coetzee M, Fontenille D. Species identification within the Anopheles funestus group of malaria vectors in Cameroon and evidence for a new species. Am J Trop Med Hyg. 2003;69:200–205. [PubMed] [Google Scholar]
- Wilkes TJ, Matola YG, Charlwood JD. Anopheles rivulorum, a vector of human malaria in Africa. Med Vet Entomol. 1996;10:108–110. doi: 10.1111/j.1365-2915.1996.tb00092.x. [DOI] [PubMed] [Google Scholar]
- De Meillon B, Van Eeden GJ, Coetzee L, Coetzee M, Meiswinkel R, Du ToitLN, Hansford CF. Observations on a species of the Anopheles funestus subgroup, a suspected exophilic vectorof malaria parasites in northeastern Transvaal, South Africa. Mosq News. 1977;37:657–661. [Google Scholar]
- Hargreaves K, Koekemoer LL, Brooke BD, Hunt RH, Mthembu J, Coetzee M. Anopheles funestus resistant to pyrethroid insecticides in South Africa. Med Vet Entomol. 2000;14:181–189. doi: 10.1046/j.1365-2915.2000.00234.x. [DOI] [PubMed] [Google Scholar]
- Brooke BD, Kloke G, Hunt RH, Koekemoer LL, Temu EA, Taylor ME, Small G, Hemingway J, Coetzee M. Bioassay and biochemical analysis of insecticide resistance in southern African Anopheles funestus (Diptera: Culicidae) Bull Entomol Res. 2001;91:265–272. doi: 10.1079/ber2001108. [DOI] [PubMed] [Google Scholar]
- Hargreaves K, Hunt RH, Brooke BD, Mthembu J, Weeto MM, Awolola TS, Coetzee M. Anopheles arabiensis and An. quadriannulatus resistance to DDT in South Africa. Med Vet Entomol. 2003;17:546–547. doi: 10.1111/j.1365-2915.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) Test Procedures for Insecticide Resistance Monitoring in Malaria Vectors, Bio-efficacity and Persistence of Insecticides on Treated Surfaces Document WHO/CDS/CPC/MAL/9812. WHO, Geneva, Switzerland; 1998. [Google Scholar]
- Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction (PCR) assay to identify the Anopheles funestus (Diptera: Culucidae) group. Am J Trop Med Hyg. 2002;36:125–136. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- Wirtz RA, Zavala F, Charoenvit Y, Campbell GH, Burkot TR, Schneider I, Esser KM, Beaudoin RL, Andre RG. Comparative testing of Plasmodium falciparum sporozoite monoclonal antibodies for ELISAdevelopment. Bull World Health Organ. 1987;65:39–45. [PMC free article] [PubMed] [Google Scholar]
- Snounou G, Viriyakoso S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Beier JC, Perkins PV, Wirtz RA, Koros J, Diggs D, Gargan TPII, Koech LK. Blood- meal identification by direct enzyme-linked immunosorbent assay (ELISA), tested on Anopheles (Diptera: Culicidae) in Kenya. J Med Entomol. 1988;25:9–16. doi: 10.1093/jmedent/25.1.9. [DOI] [PubMed] [Google Scholar]
- Penilla RP, Rodríguez AD, Hemingway J, Torres JL, Arredondo-Jiménez JI, Rodríguez MH. Resistance management strategies in malaria vector mosquito control. Baseline data for large-scale field trial against Anopheles albimanus in Mexico. Med Vet Entomol. 1998;12:217–233. doi: 10.1046/j.1365-2915.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- Kamau L, Koekemoer LL, Hunt RH, Coetzee M. Anopheles parensis : the main member of the Anopheles funestus species groupfound resting inside humans dwellings in Mwea area of central Kenyatoward the end of the rainy season. J Am Mosq Contr Assoc. 2003;19:130–133. [PubMed] [Google Scholar]
- Koekemoer LL, Rankoe EM, La Grange JP, Govere J, Coetzee M. False detection of Plasmodium falciparum sporozoites in Anopheles marshallii group mosquitoes. J Am Mosq Contr Assoc. 2001;17:160–165. [PubMed] [Google Scholar]
- Lochouarn L, Fontenille D. ELISA detection of malaria sporozoites: false-positive results in Anopheles gambiae s.l. associated with bovine blood meals. Trans R Soc Trop Med Hyg. 1999;93:101–102. doi: 10.1016/S0035-9203(99)90198-0. [DOI] [PubMed] [Google Scholar]
- Somboon MN, Koottathep S, Trisanarom U. Detection of sporozoites of Plasmodium vivax and Plasmodium falciparum associated with bovine and swine blood. Trans R Soc Trop MedHyg. 1993;87:322–324. doi: 10.1016/0035-9203(93)90148-J. [DOI] [PubMed] [Google Scholar]
- Swellengrebel NH, De Buck A. Malaria in the Netherlands. Scheltema and Holkema; Amsterdam Ltd; 1938. [Google Scholar]