Abstract
Ewing sarcoma gene EWS encodes a putative RNA-binding protein with proposed roles in transcription and splicing, but its physiological role in vivo remains undefined. Here, we have generated Ews-deficient mice and demonstrated that EWS is required for the completion of B cell development and meiosis. Analysis of Ews–/– lymphocytes revealed a cell-autonomous defect in precursor B lymphocyte (pre–B lymphocyte) development. During meiosis, Ews-null spermatocytes were deficient in XY bivalent formation and showed reduced meiotic recombination, resulting in massive apoptosis and complete arrest in gamete maturation. Inactivation of Ews in mouse embryonic fibroblasts resulted in premature cellular senescence, and the mutant animals showed hypersensitivity to ionizing radiation. Finally, we showed that EWS interacts with lamin A/C and that loss of EWS results in a reduced lamin A/C expression. Our findings reveal essential functions for EWS in pre–B cell development and meiosis, with proposed roles in DNA pairing and recombination/repair mechanisms. Furthermore, we demonstrate a novel role of EWS in cellular senescence, possibly through its interaction and modulation of lamin A/C.
Introduction
Ewing sarcoma gene EWS was first identified from the Ewing sarcoma chromosomal breakpoint t(11;22)(q24;q12) region as a translocation-generated fusion gene product between EWS and FLI1, a member of the ETS family of transcription factors (1). It was soon discovered that EWS was frequently rearranged in chromosomal translocations in other types of cancer and invariably led to a fusion of EWS to various transcription factor genes (reviewed in refs. 2, 3). The transcription factor partners include ATF1, a bZIP transcription factor, in malignant melanoma of soft parts (4); C/EBP-homologous protein (CHOP) in myxoid liposarcoma (5); Wilms tumor gene 1 (WT1) in desmoplastic small round cell tumor (DSRCT) (6); and NR4A3/CHN/NOR-1 in extraskeletal myxoid chondrosarcoma (7). In all cases, the aminoterminal domain (NTD) of EWS is fused, in frame, to a DNA-binding domain of various transcription factors, creating an aberrant transcription factor that is not present in normal cells. It is thus likely that the different pathophysiology of various EWS-fusion driven sarcomas is largely determined by a particular DNA-binding domain encoded by the EWS translocation partner.
EWS is a member of the TET family of proteins, which includes 3 other members, translocated in liposarcoma/fusion (TLS/FUS), TAF15/hTAFII68 (TATA-binding protein–associated factor 15/human TATA-binding protein–associated factor II68), and the Drosophila protein cabeza/SARFH, all of which contain a putative RNA-binding domain (8). The NTDs of EWS, TLS, and TAF15 contain degenerate repeats of the SYGQ motif and mediate potent transcriptional activation when fused to heterologous DNA-binding domains of different transcription factors (9–11). This domain is also essential for the transforming activity of EWS/Fli1 (12). Interestingly, the NTDs of EWS and TLS appear to be functionally interchangeable, as chromosomal translocation of either EWS-CHOP or TLS-CHOP is found in myxoid liposarcoma, with an indistinguishable pathophysiology (10, 13). Similarly, in extraskeletal myxoid chondrosarcoma, rearrangements of either EWS or TAF15 NTD to NR4A3 transcription factor have been found (7, 14, 15). The carboxyterminal domain (CTD) of EWS contains an RNA recognition motif (RRM), 3 RGG box domains, and a Ran-binding protein 2–like (RanBP2-like) zinc finger motif (1). All the TET proteins have been shown to bind RNA in vitro in a nonspecific manner as well as single-strand DNA (ssDNA) (8, 16–18). Surprisingly, in vitro RNA binding by EWS only required the RGG box and not the RRM domain (16). In addition, EWS contains an IQ domain that is phosphorylated by PKC and is implicated in binding to calmodulin (19). Interestingly, PKC phosphorylation of EWS or EWS-calmodulin interaction led to a decrease in RNA binding by EWS, suggesting a possible regulatory mechanism for EWS-RNA interaction.
EWS has been shown to interact with subunits of transcription factor II D and RNA polymerase II complexes (20) and various splicing factors (21, 22), but its role in basal transcription or splicing has yet to be demonstrated. A transcriptional role of EWS has been inferred from studies that demonstrated EWS interaction with RNA polymerase II, TFIID, and CBP (23); subsequently, EWS was shown to influence transcriptional activities of HNF3 (24), BRN3A (25), and OCT4 (26) in reporter assays. A role of EWS in splicing was suggested from its interactions with various splicing factors, SF1/ZFM1 (27), U1C (21), YB1 (28), and TASR-1 and -2 (22). However, EWS did not show any effects on splicing in in vitro splicing assays, while EWS-Fli1 inhibited U1C-, YB1-, and TASR-mediated splicing (21, 22, 28). To determine the in vivo role of EWS, we used gene targeting in mouse ES cells to inactivate EWS in mice. Our analysis of Ews-deficient mice revealed that EWS is essential for precursor B lymphocyte (pre–B lymphocyte) development and meiosis. Furthermore, studies with mouse embryonic fibroblasts (MEFs) demonstrated an involvement of EWS in cellular senescence, possibly through its interaction with nuclear lamin A/C.
Results
Inactivation of Ews in mouse ES cells.
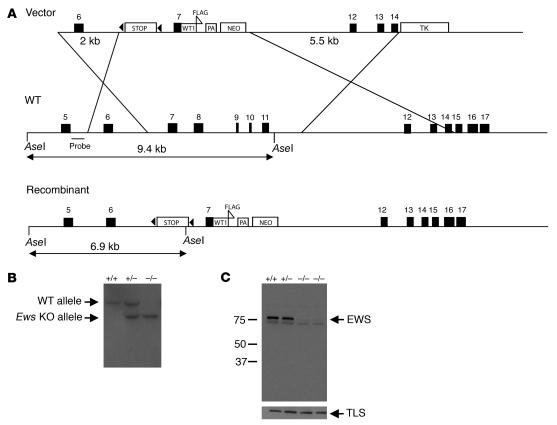
During the generation of conditional knock-in mice expressing the EWS/WT1 translocation gene product, we generated an Ews-null allele by inserting a stop sequence flanked by 2 loxP sequences (29) in intron 6 of Ews (Figure 1A). In addition, Ews exons 8–11, encoding a part of the RRM motif, were deleted by homologous recombination (Figure 1A). Targeted mouse ES clones were identified and used to generate chimeras, which were subsequently bred with Black Swiss outbred mice to establish F1 heterozygotes (Ews+/tmStop, designated herein as Ews+/–) mice. Heterozygous Ews+/– mice were intercrossed to generate Ews–/– animals (Figure 1B). To determine whether the insertion of stop resulted in a complete loss of EWS, we examined EWS expression in MEFs by Western blot analysis using rabbit polyclonal antibodies raised against the first 74 amino acids of mouse Ews. As shown in Figure 1C, EWS expression was completely absent in the Ews–/– cells. A truncated EWS was not detectable even after a long exposure of the immunoblot. In addition, loss of EWS did not result in a compensatory increase in the expression of its homolog TLS/FUS (Figure 1C). Northern blot analysis revealed a complete absence of full-length Ews transcripts but an accumulation of short truncated transcripts in Ews-null MEFs (data not shown). Although the possibility remains that an undetectable amount of truncated Ews protein may exist, Ews-heterozygous mice were indistinguishable from wild-type littermates and developed normally, suggesting that the targeting strategy resulted in an Ews-null allele.
Figure 1. Inactivation of Ews by gene targeting.
(A) Scheme for Ews targeting strategy. The triangles indicate loxP sequences flanking the stop cassette. WT1, WT1 cDNA; PA, poly(A) sequence. The probe used for Southern blot analysis is indicated. (B) Southern blot analysis of AseI-digested tail DNA. Wild-type (9.4-kb) and targeted (6.9-kb) alleles are marked. (C) Western blot analysis of EWS and TLS using whole-cell extracts isolated from MEFs. Ews genotype and the molecular weight markers (kDa) are indicated.
Ews-deficient mice are runted and show high postnatal lethality.
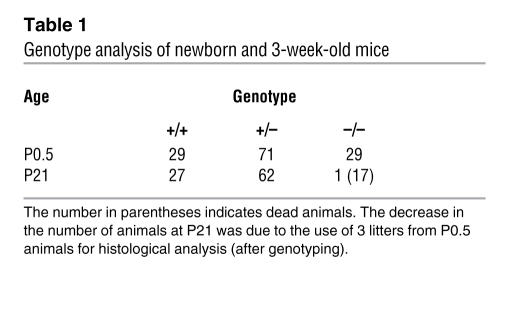
EWS is a ubiquitously expressed protein, and based on its predicted functions in basal transcription and splicing (3), we presumed that a complete absence of Ews would interfere with normal development. Surprisingly, Ews-null animals were born at the expected Mendelian ratio (Table 1). However, all of the homozygous mutants were born smaller than their littermates and remained small through weaning (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI31222DS1). We also observed a high rate (about 90%) of postnatal mortality in Ews–/– pups prior to weaning. A small number of the mutant animals did not suckle and may have died from lack of feeding. However, most Ews mutant pups were found dead or disappeared despite evidence of feeding. Examination of early postnatal Ews mutant pups revealed normal appearance of all major organs; therefore, the exact cause of death is not known. We obtained a few surviving Ews–/– animals for further characterization. It is worth noting that we did not get any surviving homozygotes in 129SvEv or C57BL/6 inbred mice.
Table 1 .
Genotype analysis of newborn and 3-week-old mice
Severe lymphopenia and a cell-autonomous pre–B cell development defect in EWS mutant mice.
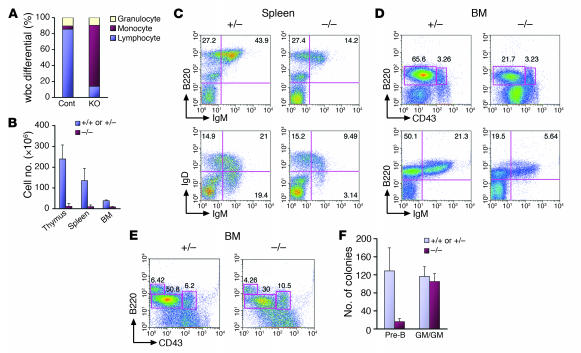
Peripheral blood analysis of 3- to 4-week-old EWS mutant animals compared with littermate controls revealed severe lymphopenia (Figure 2A). Erythropoiesis seemed to be normal, since both wild-type and mutant mice showed similar red blood cell count and hematocrits (data not shown). All of the Ews-null animals examined displayed disproportionately small thymi and spleens compared with their littermates, and this was reflected by the markedly reduced cellularity of these organs (Figure 2B). To examine the lymphocyte development in detail, we performed flow cytometric analysis of T and B lymphocytes. Flow cytometry of T cells in thymus indicated only a slight decrease in the proportion of CD4+CD8+ cells in the mutants compared with controls (Supplemental Figure 2). Further analysis of CD4–CD8– precursor T cells with CD25 and CD44 markers also revealed no major alterations. However, analysis of B lymphocytes revealed marked reductions in both immature (B220+IgMhi) and more mature (IgMhiIgDhi) B cell populations from bone marrow and spleen of EWS mutant animals (Figure 2, C and D), suggesting a developmental defect in B lymphocyte maturation. Flow cytometry using markers to identify pre–B lymphocytes (B220+CD43–) from the Ews–/– bone marrow demonstrated a marked reduction in the pre–B cell compartment (Figure 2D). In contrast, progenitor B cell (pro–B cell) populations (B220+CD43+) appeared to be normal. These results suggest that there might be a developmental arrest in the pro–B to pre–B cell transition in the absence of EWS. To confirm this observation, we performed an in vitro pre-B CFU assay using bone marrow cells isolated from wild-type and mutant animals. The results showed that progenitor cells isolated from Ews–/– bone marrow failed to form pre-B colonies in vitro but differentiated as efficiently into other lineages (GM-CFU) as the control (data not shown; also see Figure 2F).
Figure 2. EWS is intrinsically required for pre–B cell development.
(A) A complete blood count was performed with peripheral blood from 3-week-old littermates (n = 3/genotype). White blood cell differential data indicate a marked reduction of the lymphocyte population in Ews–/– mice. (B) Reduced cellularity of lymphoid organs in Ews–/– mice. Total cell count is shown for indicated organs of 3-week-old mice (n = 3/genotype). Flow cytometry of splenocytes (C) and bone marrow–derived cells (D) from Ews–/– mice and littermate controls using B cell markers B220, CD43, IgM, and IgD. (E) Fetal liver chimera analysis. Bone marrow cells were harvested from fetal liver chimeras and analyzed by flow cytometry with antibodies against CD43 and B220 along with CD45.1 and CD45.2. Presented are representative data from 3 independent experiments with similar results (n = 3 for Ews–/– and n = 3 for Ews+/– littermate controls). (F) CFU assay. Bone marrow cells from fetal liver chimeras were plated in duplicate in either pre-B or multilineage differentiation (GM-CFU) media. The number of colonies is presented as the mean colony number from 3 independent experiments.
To further demonstrate the intrinsic nature of the pre–B cell defect in vivo, we performed a fetal liver transplantation experiment. To this end, lethally irradiated Rag2–/– hosts (B6.SJL strain, CD45.1+) were reconstituted with E14.5 fetal liver–derived hematopoietic precursors of all 3 Ews genotypes (129/Black Swiss hybrids, CD45.2+) by tail-vein injection and analyzed 6 weeks later by flow cytometry. Analysis of donor-derived T lymphocytes revealed a relatively normal reconstitution of all T cells by the EWS mutant fetal liver cells (Supplemental Figure 2C). However, reconstitution of the pre–B cell population (B220+CD43–) was markedly reduced in animals injected with the EWS mutant cells, whereas the B220+CD43+ pro–B cell population appeared normal compared with that of control (Figure 2E). Consistent with the results of our pre-B colony assay, bone marrow progenitors reconstituted from Ews–/– fetal liver cells were unable to differentiate into pre-B colonies in vitro but developed efficiently into myeloid/granulocytic lineages (Figure 2F). Together, these results clearly demonstrate that EWS is intrinsically required for pre–B lymphocyte development.
Homozygous inactivation of Ews leads to meiotic arrest and apoptosis.
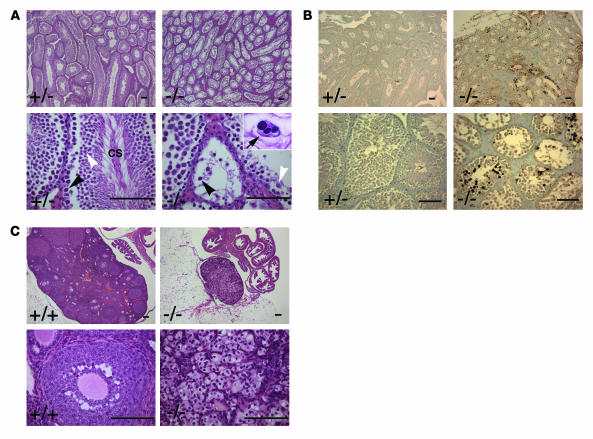
Homozygous EWS mutant male and female mice failed to produce any pups when mated with wild-type animals. Upon gross examination, testis and ovary were found to be greatly reduced in size in all EWS mutant animals examined (data not shown). Histologic analysis of testes from 12-week-old EWS mutants revealed a complete maturation arrest of spermatids (Figure 3A). The seminiferous tubule contained primary spermatocytes, but there were no condensing spermatids in the seminiferous tubules or any maturing spermatozoa in the epididymis. Spermatogonia were present, but primary spermatocytes and round spermatids were disorganized and markedly reduced in number. In Ews–/– testes, we also observed spermatocytes that were enlarged and multinucleated, possibly as a result of abnormal segregation of meiotic chromosomes and cell division (Figure 3A, inset, arrow). The wild-type testes contained normal primary spermatocytes, round spermatids, and condensing spermatids but did not contain any abnormal multinucleated spermatocytes. These results suggest that the Ews–/– primary spermatocytes were undergoing spermatic arrest at the point of or just prior to the first meiotic division. Consistent with this, TUNEL staining showed the occurrence of massive apoptosis of primary spermatocytes in the EWS mutant testes, but TUNEL-positive apoptotic spermatocytes were rarely found in the controls (Figure 3B).
Figure 3. Ews–/– mice are defective in spermatogenesis and oogenesis.
(A) H&E staining of testes from 3-month-old Ews–/– and Ews+/– littermates. Black arrowheads mark primary spermatocytes, and white arrowheads indicate round spermatids. CS, condensing spermatids. The arrow in the inset shows a multinucleated spermatocyte. (B) TUNEL staining of testes from 3-month-old Ews–/– and Ews+/– littermates shows massive apoptosis of spermatocytes in the mutant testes. (C) H&E staining of ovaries from 3-month-old Ews–/– and Ews+/– littermates. Note the complete absence of oocytes and developing follicles in Ews–/– ovary (right panels). A minimum of 3 animals for each genotype were examined for H&E and TUNEL analyses. Scale bars: 100 μm.
Twelve-week-old female homozygous mice were devoid of maturing ovarian follicles and showed mild proliferation of interstitial cells (Figure 3C). There were no corpora lutea present in the ovaries of most Ews–/– female mice examined. In younger Ews-null female mice (3 weeks), however, we observed immature ovarian follicles, suggesting that the defects probably occurred during the later stages of oocyte maturation (data not shown). These results demonstrate that EWS is essential for gametogenesis in both sexes.
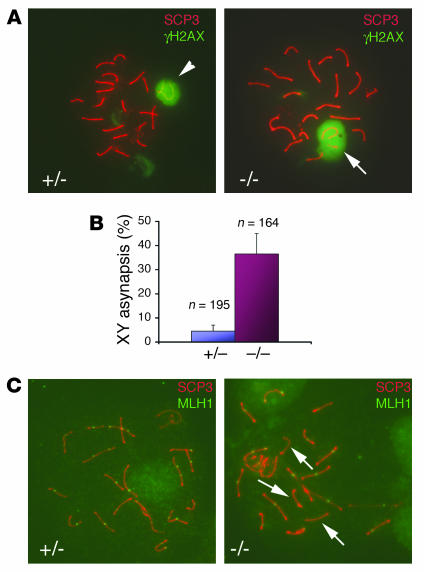
Ews deficiency leads to XY asynapsis and reduction in meiotic recombination.
Meiotic prophase I in spermatogenesis proceeds sequentially in morphologically distinguishable stages: leptotene, zygotene, pachytene, diplotene, and diakinesis (30). Each stage can be distinguished by immunostaining of synaptonemal proteins (e.g., synaptonemal complex protein 3 [SCP3]) present at the synaptonemal complex formed between homologous chromosomes (31). To examine the spermatogenesis defects in Ews-null mice in detail, we analyzed spermatocytes from 4- to 6-week-old mice by immunostaining of meiotic chromosome spreads with antibodies against SCP3 and γ-H2AX, which is a marker of the sex body containing XY bivalent in pachynema (32). During the pachytene stage, sex chromosomes form a partial synapsis through their respective pseudoautosomal regions (Figure 4A, arrowhead). However, in Ews–/– spermatocytes, more than one-third of pachytene nuclei (60 of 164) showed asynapsis of XY chromosomes (Figure 4A, arrow). In comparison, only 9 of 195 nuclei from Ews+/– spermatocytes contained univalent XY (Figure 4B).
Figure 4. Reduced meiotic recombination and XY asynapsis in Ews–/– spermatocytes.
(A) Double immunofluorescence staining of spermatocyte spread from 6-week-old testes with α-SCP3 and α-γH2AX antibodies. The arrowhead (left panel) shows a sex body containing bivalent XY, and the arrow (right panel) indicates univalent XY. Original magnification, ×40. (B) Univalent XY were counted under a fluorescence microscope and represented as the percentage of total pachytene nuclei examined (3 mice/genotype). n values represent total pachytene nuclei. (C) Double immunofluorescence staining of spermatocyte spread with α-SCP3 and α-MLH1 antibodies. The arrows in the right panel indicate bivalents with no MLH1 staining. Original magnification, ×40.
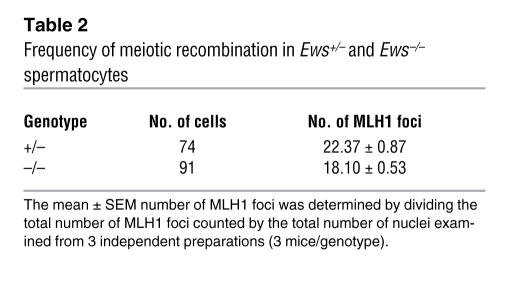
After the formation of bivalents, synapsed chromosomes undergo programmed double-strand breaks (DSBs) and initiate meiotic recombination by homologous recombination. Meiotic recombination is essential for the proper segregation of chromosomes during meiosis (30). Meiotic crossovers, which are sites of DSB and homologous recombination, can be visualized by immunostaining with an antibody against MLH1 (33), a homolog of the E. coli DNA mismatch repair gene mutL. To determine whether EWS mutant spermatocytes undergo normal meiotic recombination, we doubled stained spermatocyte nuclei with antibodies against SCP3 and MLH1. In control pachytene nuclei, all bivalents contained at least 1 MLH1 foci, with an average of 22 MLH1 foci per genome (Figure 4C, left panel, and Table 2); this is consistent with the approximately 23 MLH1 foci per genome present in normal mouse spermatocytes (30). In contrast, there was a reduction in the number of MLH1 foci in the absence of EWS compared with the control (Figure 4C, right panel, and Table 2). Furthermore, MLH1 staining was conspicuously absent in some of the bivalents in the Ews–/– pachynema (Figure 4C, arrows, right panel), suggesting that the meiotic recombination had not occurred. These results demonstrate that EWS is critically important in meiosis, specifically in the formation of XY bivalent and meiotic crossovers.
Table 2 .
Frequency of meiotic recombination in Ews+/– and Ews–/– spermatocytes
Loss of EWS leads to premature cellular senescence and hypersensitivity to ionizing radiation.
We next investigated the cellular effects of EWS deficiency. Thus, we generated Ews-null and litter-matched control MEFs and examined their cellular properties. During the early passages, Ews-null MEFs grew slightly slower than the control MEFs, but the proliferation soon ceased, after passages 3–4, whereas the control cells continued to grow to passage 5 and beyond (Figure 5A). Ews-deficient MEFs at passage 3 readily displayed endogenous senescence-associated β-galactosidase activity, a marker of cellular senescence (34), which was rarely seen in the passage 3 control cells (Figure 5, B and C). Two major pathways, p53 and retinoblastoma (RB), are considered to be important in cellular senescence (35). Thus, we examined whether the senescence observed in Ews-null cells was associated with increased expression of p53, p21Cip1, p19Arf, or p16Ink4a. Immunoblot analysis demonstrated that the expression levels of these proteins remain relatively unchanged in the mutant cells undergoing senescence (passage 4) as compared with controls (Figure 5D). Interestingly, the expression level of pRb was reduced in Ews–/– cells compared with the control MEFs. The reduced pRb expression, however, is more likely to result in cell proliferation than senescence, as the acute loss of pRb in mouse fibroblasts has been shown to override cellular senescence (36).
Figure 5. Premature cellular senescence and aging-like features in Ews–/– mice.
(A) MEFs were continuously passaged, and cell count determined at every passage is plotted. n = 3/genotype. (B) Senescence-associated β-galactosidase staining of passage 3 Ews+/+ and Ews–/– MEFs. A representative field is shown. Original magnification, ×20. (C) Cells positively stained for senescence-associated β-galactosidase activity were counted under a microscope. Results represent an average of 4 independent cell lines examined at passage 3 for each genotype. (D) Western blot analysis of p19, p21, p53, p16, pRb, EWS, and actin using 3 independently derived MEFs of each genotype. (E) Representative x-ray images of the whole body and the dissected long bones of littermates are shown. X-rays were taken of 3- to 6-week-old mice of the indicated genotypes (n = 4). (F) H&E staining of skin sections from a similar dorsal area of 3-week-old littermates. F, subcutaneous fat. M, muscle. Scale bars: 100 μm. (G) Kaplan-Meier survival curve of mice that received 7 Gy of ionizing radiation on day 1.
Cellular senescence observed in Ews-null cells was accompanied by an aging-like phenotype in Ews-deficient mice. In EWS mutant mice, we observed discernible aging-like characteristics (37), such as kyphosis (Figure 5E, left panel), reduced bone density (Figure 5E, right panel), and loss of subcutaneous fat and the atrophy of accompanying muscle in skin (Figure 5F), which were not evident in the littermate controls. The degree of kyphosis was variable in the mutant animals, but even in Ews-null mice with less severe kyphosis, the mutant animals displayed reduction in the thickness of the subcutaneous adipose layer and reduction in bone density (data not shown). Accelerated aging is frequently seen in mice with defects in the DNA damage repair pathway (38). Therefore, we examined whether Ews–/– animals may be hypersensitive to γ-radiation. Our result demonstrated that Ews-deficient mice are highly susceptible to ionizing radiation (Figure 5G). Most of the EWS mutant mice (6 of 7) irradiated at 7 Gy died or were moribund by 60 days, whereas 8 of 9 littermate controls (Ews+/+ and Ews+/–) survived beyond 120 days. We observed a similar hypersensitivity to various types of DNA damage in Ews-null MEFs compared with MEFs derived from litter-matched controls (Supplemental Figure 3).
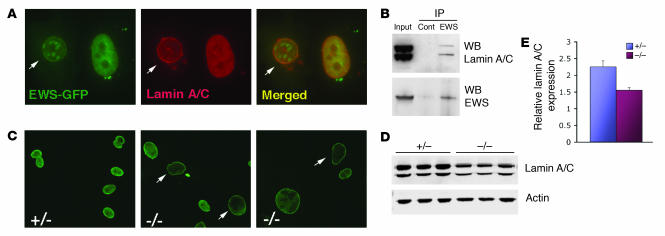
EWS physically associates with nuclear lamin A/C.
Recent reports have shown that mutations in LMNA, which encodes A-type lamins, are responsible for a variety of diseases collectively termed laminopathies (reviewed in ref. 39), including Hutchinson-Gilford progeria syndrome (HGPS) (40, 41). The observed aging-like features and asynapsis of XY in Ews-null mice are characteristics shared by Lmna–/– mice (42, 43). Therefore, we considered a possible link between EWS and lamin A/C by first examining their subcellular localization. Cells transfected with GFP-EWS fusion constructs showed mostly diffuse expression of GFP-EWS in the nucleoplasm, and although rare, some cells also showed colocalization of GFP-EWS with endogenous lamin A/C in the perinuclear region (Figure 6A, arrow). GFP-EWS was also found in punctate nuclear structures, which did not colocalize with lamin A/C. Lamin A/C was expressed in both perinuclear and intranuclear regions (Figure 6A). To demonstrate a direct in vivo association between EWS and lamin A/C, endogenous EWS was immunoprecipitated from HeLa nuclear extracts, and the presence of lamin A/C in the complex was examined by immunoblotting. As shown in Figure 6B, endogenous EWS and lamin A/C physically associated in vivo. We next examined the expression of endogenous lamin A/C in Ews-null MEFs. Ews-deficient cells contained enlarged nuclei and showed markedly reduced intranuclear staining of lamin A/C compared with controls (Figure 6C, arrows). There was a consistent reduction in lamin A/C expression in all independently derived Ews-null fibroblasts compared with MEFs derived from litter-matched controls (Figure 6, D and E). The level of lamin A/C transcripts was similar in both control and mutant MEFs (data not shown), demonstrating that inactivation of EWS results in a reduction of lamin A/C at a posttranscriptional level.
Figure 6. Endogenous interaction of EWS and lamin A/C.
(A) Localization of GFP-EWS and endogenous lamin A/C in PC3 cells by immunofluorescence microscopy. Arrow in the merged panel indicates the colocalization (yellow) in the inner nuclear membrane. Original magnification, ×40. (B) HeLa nuclear extracts immunoprecipitated with either preimmune (control) or EWS antibodies were immunoblotted with α-lamin A/C antibodies. WB, Western blot. (C) Immunofluorescence microscopy of endogenous lamin A/C in Ews+/– or Ews–/– MEFs. Original magnification, ×20. (D) Western blot analysis of MEFs with indicated genotypes using α-lamin A/C or α-actin antibodies. (E) Quantification of lamin A/C relative to actin in the immunoblot (D) using the Odyssey infrared system.
Discussion
Ubiquitous RNA-binding protein EWS has been suggested to play fundamental roles in both transcription and splicing and is thus presumed to be essential for normal development. In the present study, we show that EWS is dispensable for embryonic development. Our characterization of Ews-null mice demonstrates that EWS is essential for pre–B lymphocyte development and meiosis. A severe reduction in cellularity of thymus in the EWS mutant animals also suggests a thymic development defect. Yet the relatively normal ratios of thymic subpopulations in the Ews–/– and Ews-null fetal liver–transplanted animals (Supplemental Figure 2) suggest that the reduction in thymic cellularity may be due to stromal or other defects. Furthermore, we have uncovered an unexpected role for EWS in cellular senescence, possibly through its interaction with nuclear lamin A/C. Although our results do not preclude a role for EWS in the fundamental processes of transcription and splicing, as other TET proteins could serve compensatory roles, our work clearly demonstrates that EWS has unique functions that other TET proteins cannot compensate.
EWS has essential and nonredundant functions during B lymphocyte development and meiosis.
The members of the TET family, EWS, TLS/FUS, and TAF15/hTAFII68, are highly related proteins that can bind RNA as well as ssDNA in vitro (3). Evidence from human cancer studies suggests that at least the NTDs of EWS, TLS, and TAF15 are functionally interchangeable (5, 7, 14, 15, 44, 45). However, our analyses of Ews–/– mice clearly demonstrate a lack of functional redundancy between EWS and TLS. Similar to deletion of Ews, inactivation of Tls in mice leads to defects in B cell development (46), but the molecular details of EWS and TLS deficiencies are different. The B cell defect in Tls-null mouse is non–cell autonomous as demonstrated by fetal liver reconstitution experiments in which the B cell populations were fully reconstituted by the Tls-null donor cells (46). The reconstituted Tls–/– mature B cells, however, are dysfunctional in the primary antibody production in response to antigen stimulation. In contrast, Ews deficiency results in an intrinsic defect in pre–B cell development, specifically during the pro–B to pre–B cell transition, as demonstrated by flow cytometry, in vitro pre-B colony formation assay, and fetal liver chimera analysis (Figure 2).
The functional differences between EWS and TLS are also evident during meiosis. Ews-null spermatocytes show a high frequency of unsynapsed XY chromosomes but show normal formation of autosomal bivalents (Figure 4). In contrast, Tls-deficient spermatocytes display defects in the formation of autosomal bivalents (47). Expression of EWS is diffuse throughout spermatocytes (Supplemental Figure 4), while TLS expression is excluded from the sex body region (47). This lack of TLS expression in the sex body provides a rationale for the XY asynapsis observed in Ews-null mice. The disruption of bivalent formation in meiosis further suggests that EWS and TLS may participate in the annealing of homologous DNA sequences, with TLS playing a major role in the formation of autosomal bivalents and EWS participating in the formation of XY bivalent. Consistent with this notion, TLS has been shown to promote homologous DNA annealing in vitro (18), and Tls-deficient cells lack this in vitro DNA pairing activity (47). Thus, our results are consistent with a role for EWS in promoting homologous DNA pairing during the formation of XY bivalent. During the preparation of this manuscript, Guipaud et al. (48) showed that, as we proposed, EWS possesses homologous DNA pairing activity in vitro.
Another meiotic defect observed in the Ews–/– spermatocytes is a reduced number of meiotic crossovers (Figure 4 and Table 2), which has not been reported in the Tls–/– mice. Decreased meiotic crossovers in Ews-null spermatocytes may be an indirect consequence of XY asynapsis. However, we observed reduced crossovers even when a normal XY bivalent was present. Furthermore, some bivalents showed a complete absence of crossovers (Figure 4C), suggesting that EWS may play a direct role in meiotic recombination. We also observed an aberrant segregation of chromosomes in Ews–/– spermatocytes (Figure 3A, arrow). It has been well established that absence or reduced numbers of crossovers can lead to improper segregation of homologous chromosomes and arrest in meiosis (30). Thus, reduction in crossover formation and XY asynapsis probably result in the massive apoptosis and meiotic arrest observed in Ews-deficient spermatocytes. Finally, loss of EWS leads to infertility in both sexes, but the TLS deficiency results only in male sterility (47). These observations demonstrate that despite sharing highly homologous sequences and the same interacting proteins (such as TFIID and RNA polymerase II), EWS and TLS serve nonredundant functions in vivo during B lymphocyte development and meiosis.
A role for the TET family of proteins in DNA recombination or repair?
Ews deficiency in mice leads to developmental defects in meiosis and pre–B cell formation. Pleiotropic defects in lymphocyte development, meiosis, and cellular senescence (or aging) are features also observed in mice deficient in Atm and c-abl (reviewed in ref. 38), 2 genes that are critical to the DNA damage/repair pathway. These observations implicate a possible role of EWS in the DNA recombination and/or repair process. Consistent with this, Ews-null mice and MEFs showed increased sensitivity to DNA damage (Figure 5G and Supplemental Figure 3). Similarly, Tls–/– mice are also radiosensitive (47), and Tls–/– MEFs display genomic instability (46). In vitro, both EWS and TLS can promote annealing of homologous DNA (18, 48), which are essential steps in double-stranded DNA break repair. Although Ews–/– MEFs do not display gross chromosomal abnormality (data not shown), our observations point to a possible role for EWS in DNA recombination and/or repair, as judged by XY asynapsis and reduction in meiotic recombination in Ews-null spermatocytes. In this regard, we note that another report demonstrated interaction of EWS with BARD1 (49), a component of the BRCA1/BARD1 complex that performs essential DNA repair and recombination functions.
Loss of EWS leads to cellular senescence and identification of lamin A/C as an EWS-interacting protein.
We uncovered an unexpected role of EWS in cellular senescence in Ews–/– fibroblasts (Figure 5). Cellular senescence can be provoked by telomere attrition when the telomere shortening reaches a critical threshold (35). However, this is unlikely given the early onset of senescence in EWS mutant cells and the presence of long telomeres in mice. The 2 major pathways involved in cellular senescence are p53 and RB, but we did not find any alterations in the levels of p53, p21, p19ARF, and p16INK4a in Ews–/– MEFs. We did observe a marked reduction in the pRb levels in the mutant cells, but this is more likely to result in proliferation than cellular senescence. These results indicate that the cellular senescence in Ews-null cells may be independent of the p53 or RB pathways or may affect these pathways in subtle ways.
Our observations suggest another mechanism by which loss of EWS leads to early onset of cellular senescence. We provide evidence for an endogenous association of EWS with nuclear lamin A/C, an intermediate-filament protein of nuclear lamina. Nuclear lamins are located in the inner nuclear membrane and form the basis of the nuclear matrix that maintains the physical structure of the nuclear envelope as well as playing important roles in transcription, DNA replication, recombination and repair, RNA splicing, and nuclear export (39). It has been well established that the mutations in LMNA, which encodes lamin A/C, result in a wide spectrum of diseases including premature aging syndromes, HGPS (40, 41), and atypical Werner syndrome (50–52). Mice with targeted mutations in Lmna show a premature aging phenotype (42) and impaired spermatogenesis with XY asynapsis defect (43). Furthermore, a recent report has demonstrated that human fibroblasts derived from old donors as well as fibroblasts derived from HGPS show greatly reduced intranuclear lamin A/C expression compared with fibroblasts from young donors (53). We observed a strikingly similar reduction in intranuclear lamin A/C expression in Ews-null fibroblasts (Figure 6C), indicating that the early onset of senescence may be due to decreased lamin A/C expression and function. Finally, cell fractionation studies have shown that EWS is a component of nuclear matrix (19). Thus, we hypothesize that EWS inactivation causes early onset of cellular senescence by reducing lamin A/C expression, which may lead to nuclear lamina dysfunction.
In conclusion, we demonstrate that EWS is essential for B cell development and meiosis. Moreover, our findings reveal previously unanticipated roles for EWS in cellular senescence and in nuclear lamin A/C expression. The presence of EWS and TLS in the nuclear matrix (19, 54) raises an intriguing possibility that other TET family proteins, especially TLS, might also be involved in nuclear lamina function. Our findings may also be relevant to EWS-related cancers in which EWS haploinsufficiency is generated by the EWS chromosomal translocation, and the potential dominant-negative effects of the EWS-fusion products on the endogenous EWS activity may contribute to tumorigenesis.
Methods
Generation of the targeting vector and Ews-deficient mouse.
129SvEv mouse genomic library was screened to obtain Ews genomic DNA. We inserted a loxP-flanked transcriptional stop cassette (29) into Ews intron 6 (Figure 1A). A human WT1(+KTS) cDNA sequence was fused to Ews exon 7 to create EWS/WT1 knock-in allele. A 5.5-kb SalI and BglII fragment containing Ews exons 12 to 14 was used as the other targeting arm, resulting in a deletion of Ews exons 8 to 11 following homologous recombination. The targeting construct was electroporated into TC-1 mouse ES cells (55). Ganciclovir- and G418-resistant clones were isolated, screened by PCR for homologous recombination, and confirmed by Southern blot analysis with a probe derived from Ews intron 5, which was PCR amplified using the primers 5'-CCATGGCTAGTTAGTAGATAAG-3' and 5'-GGGGATACAATCAGTGACTTACG-3'. Positive ES clones were injected into C57BL/6J blastocysts, and the resulting chimeras were crossed with Black Swiss females (Taconic) to generate F1 heterozygotes, which were interbred to generate homozygous Ews–/– mice. Genotyping was performed by PCR analysis of tail DNA using 2 sets of primers: wild-type (411-bp PCR product): 5'-TGGATCCTACAGCCAGGCTCC-3', 5'-TGCTCGCTAGTGCTCTGTGAGCAGGAC-3'; mutant (237-bp PCR product): 5'-TGGATCCTACAGCCAAGCTCC-3', 5'-CCTGTATGAGTCCTGGTGTGGGTC-3' (40 cycles of 94°C for 45 seconds, 58°C for 45 seconds, and 72°C for 45 seconds). Animal care and use was approved by the NIH Animal Research Advisory Committee.
Generation of rabbit polyclonal antibodies against EWS.
Two N-terminal regions of mouse Ews were amplified by PCR and subcloned into pGEX3X (Amersham) to generate GST-EWS fusion proteins containing either residues 1–74 or 68–171 of EWS. Both GST-EWS fusion proteins were expressed in E. coli and purified using GST-Sepharose beads (Amersham). Two rabbit polyclonal antibodies were raised by immunizing rabbits with either purified GST-EWS1–74 or GST-EWS68–171 fusion proteins (Strategic Biosolutions). The specificity of crude sera and affinity-purified antibodies was verified by immunoblotting (data not shown and ref. 56).
Flow cytometry.
Thymocytes, splenocytes, and bone marrow cells were harvested from 3-week-old animals using standard methods. Red blood cells were removed using NH4Cl lysis buffer (Sigma-Aldrich). Total cell number was determined using a hemocytometer. For flow cytometry, cells were suspended in 1% BSA-PBS and incubated with anti-FcR antibody (BD Biosciences). Cells were then incubated with indicated antibodies on ice for 30 minutes and washed and analyzed by flow cytometry using a FACSCalibur (BD Biosciences). The following antibodies (BD Biosciences) were used: FITC-labeled anti-CD43, FITC-labeled anti-IgD, FITC-labeled anti-CD45.2, R-PE–labeled anti-CD45.1, PE-labeled anti-IgM, peridinin chlorophyll-a protein–labeled anti-B220, and allophycocyanin-labeled anti-IgM.
Fetal liver transplantation and in vitro CFU assays.
For fetal liver transplantation experiments, embryos were obtained at E14.5 from Ews+/– intercrosses (CD45.2+, 129SvEv/Black Swiss hybrid), and fetal liver cell suspensions were injected (5 × 106) into tail veins of irradiated Rag2–/– CD45.1+ recipient mice (B6.SJL; Taconic) housed under pathogen-free conditions with medicated water. The genotypes were determined retrospectively by PCR. Six weeks later, tissues were harvested and analyzed by flow cytometry for donor-derived CD45.2+ B and T cell lineages. For CFU assays, fetal liver–transplanted bone marrow cells were plated onto 35-mm dishes in duplicate in a semisolid methylcellulose medium containing either pre-B differentiation cocktail (M3630) or multilineage differentiation cocktail (M3534; StemCell Technologies Inc.). Cells were plated at 2 × 105 per plate for the CFU pre-B assay or at 1 × 105 per plate for the GM-CFU assay. The cultures were placed in an incubator (37°C, 5% CO2) for 1 week, and colonies were counted under a microscope.
Histological analysis and TUNEL assay.
Mouse tissues, including testes and ovaries, were isolated, fixed in 10% formalin solution (Sigma-Aldrich), embedded in paraffin, and sectioned for histology (H&E). The TUNEL assay was performed with the TdT-FragEL DNA Fragmentation Detection Kit (EMD Biosciences) following the manufacturer’s instructions.
Spermatocyte preparations and immunofluorescence microscopy.
For spermatocyte preparations, 4- to 6-week-old mice were used. Spermatocyte surface-spread preparations and immunofluorescent staining were performed as described previously (57). For double staining, mouse or rabbit antibodies against SCP3 (Abcam) were used with either rabbit antibody against γ–H2AX (TREVIGEN) or mouse antibody against MLH1 (BD Biosciences). For quantification of the XY univalent and the number of MLH1 foci in pachynema, spermatocyte preparations from 3 mice of each genotype were counted under a fluorescence microscope (Leica DMLB; Leica Microsystems).
Generation of MEFs.
MEFs were generated from E14.5 embryos using a standard protocol and cultured in DMEM supplemented with 20% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen). For growth curve analysis, 5 × 105 MEFs were seeded initially, and every 3 days, cells were counted and replated at 5 × 105 cells/plate. Cumulative cell numbers were calculated at each passage. For cellular senescence, passage 3 MEFs were seeded at a density of 5 × 105 MEFs, and 3 days later, cells were fixed and stained for the senescence-associated acidic β-galactosidase activity as described previously (34).
Bone x-ray imaging and mouse irradiation.
Mice were euthanized by CO2, and the x-ray images of whole animals or the long bones were obtained using MX-20, a Faxitron X-ray Corp. instrument. X-ray doses used were 20 kV for 100 seconds for whole-body images and 16 kV for 100 seconds for the dissected long bones. For whole-body mouse irradiation, 6- to 10 month-old animals were γ–irradiated at a dose of 7 Gy (GammaCell 1000 irradiator) and the animals were monitored for survival. Moribund animals were euthanized and were considered dead on the day of the sacrifice.
Transfection, immunoprecipitation, and immunoblot analyses.
PC3 prostate cancer cells were transfected with either pEGFP-C1 (BD Biosciences) or pEGFP-C1-EWS using Lipofectamine 2000 (Invitrogen). After 48 hours, cells were fixed and immunostained with goat anti–lamin A/C antibody (Santa Cruz Biotechnology Inc.) followed by TRITC-labeled anti-goat secondary antibody (Sigma-Aldrich). The photographs were taken using a fluorescence microscope (Leica DM LB). For immunoprecipitation analysis, HeLa nuclear extracts were precleared with protein A agarose (Millipore) and incubated with either rabbit antibody against EWS or preimmune serum for 2 hours at 4°C. The immune complexes were precipitated with protein A agarose and washed 6 times with PBS, and the eluted sample was analyzed by immunoblotting. Goat anti–lamin A/C (1:200) and rabbit anti-EWS (1:1,000) were used for Western blotting. For quantitative Western blot analysis of MEFs, the following antibodies were used: TLS (BD Biosciences), p19ARF (M-60; Santa Cruz Biotechnology Inc.), p21Cip1 (F-5; Santa Cruz Biotechnology Inc.), p53 (BD Biosciences), p16INK4a (M-156; Santa Cruz Biotechnology Inc.), RB (BD Biosciences), lamin A/C (N-18; Santa Cruz Biotechnology Inc.), and actin (Sigma-Aldrich). Quantitative Western blot analysis was performed using an Odyssey infrared system (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We thank Brian Sauer for kindly providing the pBS302 plasmid containing the stop cassette; Tadashi Yamashita for his help with mouse ES cell culture; Marina Bellani, Galina Petukhova, and Xiaoling Xu for helpful advice on spermatocyte preparation and antibodies; James Simone for help with FACS; and Lisa Beers for excellent technical assistance. We thank Rick Proia, Michael Eckhaus, Byeong-Chel Lee, Cariappa Annaiah, Laura Allende, and Cao Liu for advice and discussion. This research was supported by the Intramural Research Program of the NIDDK (to S.B. Lee and C. Deng) and by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (to J. O’Shea).
Footnotes
Nonstandard abbreviations used: CHOP, C/EBP-homologous protein; HGPS, Hutchinson-Gilford progeria syndrome; hTAFII68, human TATA-binding protein–associated factor II68; MEF, mouse embryonic fibroblast; NTD, aminoterminal domain; pre–B lymphocyte, precursor B lymphocyte; pro–B cell, progenitor B cell; RB, retinoblastoma; RRM, RNA recognition motif; SCP3, synaptonemal complex protein 3; ssDNA, single-strand DNA; TLS/FUS, translocated in liposarcoma/fusion; WT1, Wilms tumor gene 1.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:1314–1323 (2007). doi:10.1172/JCI31222
References
- 1.Delattre O., et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 2.Arvand A., Denny C.T. Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene. 2001;20:5747–5754. doi: 10.1038/sj.onc.1204598. [DOI] [PubMed] [Google Scholar]
- 3.Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene. 2005;363:1–14. doi: 10.1016/j.gene.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Zucman J., et al. EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat. Genet. 1993;4:341–345. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 5.Panagopoulos I., et al. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12:489–494. [PubMed] [Google Scholar]
- 6.Ladanyi M., Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- 7.Labelle Y., et al. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum. Mol. Genet. 1995;4:2219–2226. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- 8.Bertolotti A., Lutz Y., Heard D.J., Chambon P., Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 9.May W.A., et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol. Cell. Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinszner H., Albalat R., Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 11.Bertolotti A., Bell B., Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- 12.Lessnick S.L., Braun B.S., Denny C.T., May W.A. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423–431. [PubMed] [Google Scholar]
- 13.Dal Cin P., et al. Additional evidence of a variant translocation t(12;22) with EWS/CHOP fusion in myxoid liposarcoma: clinicopathological features. J. Pathol. 1997;182:437–441. doi: 10.1002/(SICI)1096-9896(199708)182:4<437::AID-PATH882>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 14.Panagopoulos I., et al. Fusion of the RBP56 and CHN genes in extraskeletal myxoid chondrosarcomas with translocation t(9;17)(q22;q11). Oncogene. 1999;18:7594–7598. doi: 10.1038/sj.onc.1203155. [DOI] [PubMed] [Google Scholar]
- 15.Sjogren H., Meis-Kindblom J., Kindblom L.G., Aman P., Stenman G. Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer Res. 1999;59:5064–5067. [PubMed] [Google Scholar]
- 16.Ohno T., et al. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9:3087–3097. [PubMed] [Google Scholar]
- 17.Zinszner H., Sok J., Immanuel D., Yin Y., Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell Sci. 1997;110:1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- 18.Baechtold H., et al. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 19.Deloulme J.C., Prichard L., Delattre O., Storm D.R. The prooncoprotein EWS binds calmodulin and is phosphorylated by protein kinase C through an IQ domain. J. Biol. Chem. 1997;272:27369–27377. doi: 10.1074/jbc.272.43.27369. [DOI] [PubMed] [Google Scholar]
- 20.Bertolotti A., et al. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knoop L.L., Baker S.J. The splicing factor U1C represses EWS/FLI-mediated transactivation. J. Biol. Chem. 2000;275:24865–24871. doi: 10.1074/jbc.M001661200. [DOI] [PubMed] [Google Scholar]
- 22.Yang L., Chansky H.A., Hickstein D.D. EWS.Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. . J. Biol. Chem. 2000;275:37612–37618. doi: 10.1074/jbc.M005739200. [DOI] [PubMed] [Google Scholar]
- 23.Rossow K.L., Janknecht R. The Ewing’s sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001;61:2690–2695. [PubMed] [Google Scholar]
- 24.Araya N., et al. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- 25.Gascoyne D.M., Thomas G.R., Latchman D.S. The effects of Brn-3a on neuronal differentiation and apoptosis are differentially modulated by EWS and its oncogenic derivative EWS/Fli-1. Oncogene. 2004;23:3830–3840. doi: 10.1038/sj.onc.1207497. [DOI] [PubMed] [Google Scholar]
- 26.Lee J., Rhee B.K., Bae G.Y., Han Y.M., Kim J. Stimulation of Oct-4 activity by Ewing’s sarcoma protein. Stem Cells. 2005;23:738–751. doi: 10.1634/stemcells.2004-0375. [DOI] [PubMed] [Google Scholar]
- 27.Zhang D., Paley A.J., Childs G. The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J. Biol. Chem. 1998;273:18086–18091. doi: 10.1074/jbc.273.29.18086. [DOI] [PubMed] [Google Scholar]
- 28.Chansky H.A., Hu M., Hickstein D.D., Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 2001;61:3586–3590. [PubMed] [Google Scholar]
- 29.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Meth. Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 30.Hassold T., Sherman S., Hunt P. Counting cross-overs: characterizing meiotic recombination in mammals. Hum. Mol. Genet. 2000;9:2409–2419. doi: 10.1093/hmg/9.16.2409. [DOI] [PubMed] [Google Scholar]
- 31.Moens P.B. Molecular perspectives of chromosome pairing at meiosis. Bioessays. 1994;16:101–106. doi: 10.1002/bies.950160206. [DOI] [PubMed] [Google Scholar]
- 32.Mahadevaiah S.K., et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 33.Baker S.M., et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 34.Dimri G.P., et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Sage J., Miller A.L., Perez-Mancera P.A., Wysocki J.M., Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 37.Kuro-o M., et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 38.Lombard D.B., et al. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 39.Mounkes L.C., Stewart C.L. Aging and nuclear organization: lamins and progeria. Curr. Opin. Cell Biol. 2004;16:322–327. doi: 10.1016/j.ceb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 40.De Sandre-Giovannoli A., et al. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 41.Eriksson M., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mounkes L.C., Kozlov S., Hernandez L., Sullivan T., Stewart C.L. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature. 2003;423:298–301. doi: 10.1038/nature01631. [DOI] [PubMed] [Google Scholar]
- 43.Alsheimer M., et al. Disruption of spermatogenesis in mice lacking A-type lamins. J. Cell Sci. 2004;117:1173–1178. doi: 10.1242/jcs.00975. [DOI] [PubMed] [Google Scholar]
- 44.Crozat A., Aman P., Mandahl N., Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 45.Rabbitts T.H., Forster A., Larson R., Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 46.Hicks G.G., et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 47.Kuroda M., et al. Male sterility and enhanced radiation sensitivity in TLS(–/–) mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guipaud O., et al. An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics. 2006;6:5962–5972. doi: 10.1002/pmic.200600259. [DOI] [PubMed] [Google Scholar]
- 49.Spahn L., et al. Interaction of the EWS NH2 terminus with BARD1 links the Ewing’s sarcoma gene to a common tumor suppressor pathway. Cancer Res. 2002;62:4583–4587. [PubMed] [Google Scholar]
- 50.Bonne G., Levy N. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:1585–1586; author reply, 1586. doi: 10.1016/S0140-6736(03)14761-7. [DOI] [PubMed] [Google Scholar]
- 51.Chen L., et al. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- 52.Vigouroux C., Caux F., Capeau J., Christin-Maitre S., Cohen A. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:1585–1586; author reply, 1586. doi: 10.1016/S0140-6736(03)14760-5. [DOI] [PubMed] [Google Scholar]
- 53.Scaffidi P., Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. [Google Scholar]
- 54.Meissner M., Lopato S., Gotzmann J., Sauermann G., Barta A. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell Res. 2003;283:184–195. doi: 10.1016/s0014-4827(02)00046-0. [DOI] [PubMed] [Google Scholar]
- 55.Deng C., Wynshaw-Boris A., Zhou F., Kuo A., Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 56.Toretsky J.A., et al. Oncoprotein EWS-FLI1 activity is enhanced by RNA helicase A. Cancer Res. 2006;66:5574–5581. doi: 10.1158/0008-5472.CAN-05-3293. [DOI] [PubMed] [Google Scholar]
- 57.Bellani M.A., Romanienko P.J., Cairatti D.A., Camerini-Otero R.D. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm–/– spermatocytes. . J. Cell Sci. 2005;118:3233–3245.. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.