Abstract
Cytoskeletal proteins have been implicated in the pathogenesis of cardiomyopathy, but how the cytoskeleton influences the transcriptional alterations associated with adverse cardiac remodeling remains unclear. Striated muscle activator of Rho signaling (STARS) is a muscle-specific actin-binding protein localized to the Z disc that activates serum response factor–dependent (SRF-dependent) transcription by inducing nuclear translocation of the myocardin-related SRF coactivators MRTF-A and -B. We show that STARS expression is upregulated in mouse models of cardiac hypertrophy and in failing human hearts. A conserved region of the STARS promoter containing an essential binding site for myocyte enhancer factor–2 (MEF2), a stress-responsive transcriptional activator, mediates cardiac expression of STARS, which in turn activates SRF target genes. Forced overexpression of STARS in the heart sensitizes the heart to pressure overload and calcineurin signaling, resulting in exaggerated deterioration in cardiac function in response to these hypertrophic stimuli. These findings suggest that STARS modulates the responsiveness of the heart to stress signaling by functioning as a cytoskeletal intermediary between MEF2 and SRF.
Introduction
The heart responds to biomechanical stress, tissue injury, and neurohumoral activation by hypertrophic growth, which is accompanied by an increase in cardiomyocyte size and protein content, cytoskeletal reorganization, and the expression of fetal cardiac genes (1–3). When sustained, cardiac hypertrophy can lead to cardiomyopathy and heart failure, a major cause of human morbidity and mortality. Accumulating evidence indicates a pivotal role of the cardiomyocyte cytoskeleton, including the Z disc complex, in the pathogenesis of dilated cardiomyopathy and chronic heart failure (4, 5). Cardiac cytoskeletal proteins participate not only in organization of the cytoskeleton and biomechanical force production but also in the transmission of stress signals leading to changes in cardiac gene expression and function. However, the precise molecular mechanisms by which cytoskeletal proteins modulate transcription are largely unknown.
Previously, we reported that calsarcin-1, a Z disc protein, negatively regulates the calcineurin/ nuclear factor of activated T cells (calcineurin/NFAT) pathway in cardiomyocytes (6). Others have shown that the cytoskeletal protein FHL-2 inhibits ERK2-mediated prohypertrophic signaling and serum response factor–mediated transcription (SRF-mediated transcription) (7, 8). More recently, the titin-associating complex was shown to negatively regulate SRF-mediated transcription by regulating the localization of MuRF2, a ubiquitin ligase that interacts with SRF and inhibits its activity (9). Although there is evidence linking sarcomeric proteins to cardiac hypertrophy, further studies are needed to understand how cytoskeletal proteins modulate prohypertrophic transcriptional pathways.
SRF regulates the expression of immediate early and muscle-specific genes by binding a conserved sequence [CC(A/T) 6GG] known as a CArG box or serum response element (10, 11). Numerous studies have demonstrated the involvement of SRF in cardiovascular development and induction of fetal cardiac genes during cardiac hypertrophy (12, 13). Targeted deletion of SRF in the developing heart results in lethal cardiac defects with reduced expression of cardiac-specific genes (14, 15), whereas overexpression of SRF in the postnatal heart leads to cardiomyopathy with increased fetal cardiac gene expression (16). Caspase-3 cleavage of SRF in the failing heart has also been shown to generate a dominant-negative form of the protein that may account for the depression of SRF target genes in the failing myocardium (17).
Myocardin and the myocardin-related transcription factors MRTF-A (MAL/MKL1) and MRTF-B (MKL2) constitute a family of SRF coactivators (10, 18–20). Myocardin is expressed specifically in cardiac and smooth muscle cells and is sufficient and necessary for normal expression of SRF-dependent smooth muscle genes (18, 21, 22). In contrast, MRTFs are expressed in a broad range of cell types (19) and couple actin dynamics and Rho signaling pathways to SRF by redistributing from the cytoplasm to the nucleus (23). Myocyte enhancer factor–2 (MEF2) is another member of the MADS box gene family implicated in muscle development (24–26). Mef2C-null mice show embryonic lethality due to abnormal development of the heart (27). MEF2 has also been implicated in the maintenance of the slow fiber type of skeletal muscle, in the control of striated muscle metabolism, and in pathological remodeling of the adult heart in response to stress signaling by controlling the expression of muscle-specific target genes both directly and indirectly (28–31).
Striated muscle activator of Rho signaling (STARS) is a muscle-specific actin-binding protein localized to the I band (including the Z disc) and the M line of the sarcomere (32). STARS can activate SRF-mediated transcription by inducing nuclear translocation of MRTF-A and -B (33). Here we show that STARS expression is upregulated in mouse models of cardiac hypertrophy and in myopathic human hearts. Cardiac-specific and stress-inducible expression of STARS is mediated by the proximal promoter, which contains an essential MEF2-binding site. Cardiac overexpression of STARS in vivo and in vitro increases the expression of SRF-dependent fetal cardiac genes and sensitizes the heart to biomechanical stress induced by transverse aortic banding and calcineurin signaling. These findings suggest that STARS acts as an intermediary between MEF2 and SRF, and a modulator of the cardiac stress response.
Results
Upregulation of STARS expression in cardiac hypertrophy and cardiomyopathy.
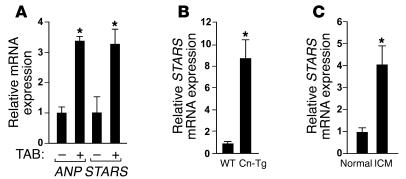
Given the changes in expression of SRF target genes during adverse cardiac remodeling and the ability of STARS to stimulate SRF activity (12, 13, 32, 33), we tested whether STARS expression might be upregulated in models of cardiac hypertrophy and cardiomyopathy. As shown in Figure 1A, STARS gene expression increased in parallel with that of atrial natriuretic peptide (ANP), a prototypical marker of cardiac remodeling, in hearts of mice subjected to pressure overload induced by thoracic aortic banding (TAB) and in hearts of Tg mice expressing constitutively active calcineurin under control of the cardiac-specific α–myosin heavy chain (α-MHC) promoter (Cn-Tg mice), a model of cardiac hypertrophy and cardiomyopathy (34) (Figure 1B). STARS expression was also significantly increased in human hearts with idiopathic cardiomyopathy (Figure 1C).
Figure 1. Upregulation of STARS during cardiac hypertrophy and failure as analyzed by real-time RT-PCR.
(A) Transcripts for STARS and ANP were detected in hearts of mice subjected to TAB and control sham operation. n = 6. (B) Expression of STARS mRNA in Cn-Tg mice and control non-Tg littermates (WT). n = 6. (C) Expression of STARS mRNA in human hearts with idiopathic cardiomyopathy (ICM) and control normal hearts. n = 3. *P < 0.05.
The STARS promoter mediates cardiac-specific and stress-inducible expression.
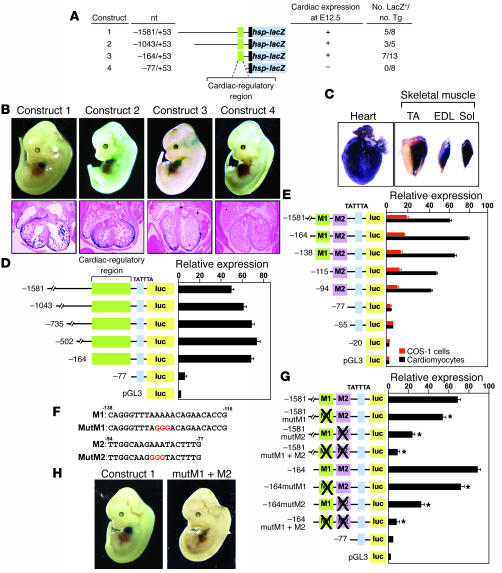
To identify the cis-regulatory elements that control STARS transcription, we generated Tg mice carrying a lacZ reporter gene controlled by various DNA fragments upstream of the mouse STARS gene. Tg mice carrying 1,581 bp of the 5'-upstream region of the STARS gene showed cardiac expression of β-galactosidase at E12.5 and in adulthood (Figure 2, A–C). The promoter also directed lacZ expression in adult skeletal muscles. Transgene expression in skeletal muscle was most prominent in the slow fiber–rich soleus muscle (Figure 2C).
Figure 2. Mutational analysis of the STARS promoter.
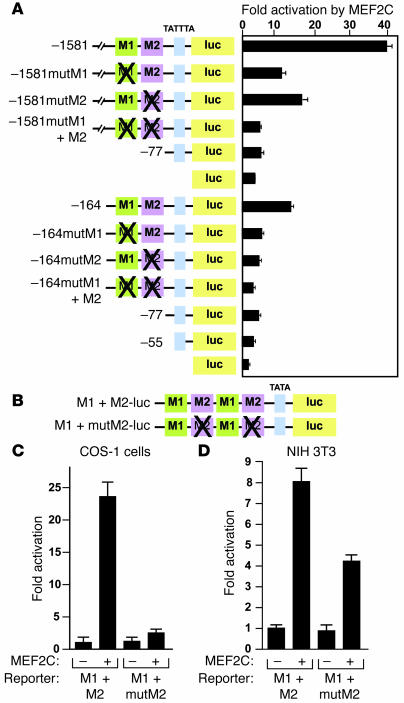
(A) Fragments of the STARS upstream region linked to the hsp68 minimal promoter and lacZ were used to generate Tg mice. (B) LacZ staining of Tg mice at E12.5 carrying the indicated STARS-lacZ transgenes. H&E-stained sections of the heart are shown. (C) LacZ staining of striated muscle of Tg mice carrying STARS-lacZ construct 1. TA, tibialis anterior; EDL, extensor digitorum longis; sol, soleus. (D and E) Luciferase activity was measured in cardiomyocytes (D and E) and COS-1 cells (E) transfected with the luciferase reporter plasmid pGL3 linked to upstream fragments of the STARS gene. (F) Sequences of the M1 and M2 regions with mutations shown in red. (G) Luciferase activity was measured in cardiomyocytes transfected with –1581- or –164STARS-luc without or with mutations in the M1 and/or M2 region (mutM1, mutM2, mutM1+M2). *P < 0.05 versus reporter without mutation. (H) LacZ staining of E12.5 Tg embryos carrying wild type construct 1 or construct 1 with mutations in the M1 and M2 regions (mutM1+M2). Original magnification, ×2 (B, upper panels, and H); ×10 (B, lower panels).
The cis-regulatory elements of the STARS upstream region were further delineated by analyzing the activity of a series of 5' deletions in Tg mice. Deletion to –164 bp upstream of the transcription initiation site did not affect cardiac-specific expression, whereas further deletion to –77 bp abolished cardiac expression of the transgene, indicating that the region between –164 bp and –77 bp is necessary for cardiac-specific expression of STARS (Figure 2, A and B).
Fragments of the STARS promoter linked to a luciferase reporter showed markedly diminished activity in cardiomyocytes when the –164 to –77 bp region was deleted (Figure 2D). Within this region, we found 2 subregions, –138 through –116 bp (M1) and –94 through –77 (M2), to mediate cardiac expression (Figure 2, E and F). A mutation in either the M1 or M2 sequences significantly reduced the cardiac activity of both –1,581-bp and –164-bp STARS promoter constructs, and a double mutation abolished promoter activity in cardiomyocytes (Figure 2G). Consistent with these findings, expression of a lacZ reporter gene controlled by the –1,581-bp STARS promoter with mutations in M1 and M2 was dramatically reduced in the hearts of Tg mice (Figure 2H), indicating that the M1 and M2 regions cooperatively mediate cardiac expression.
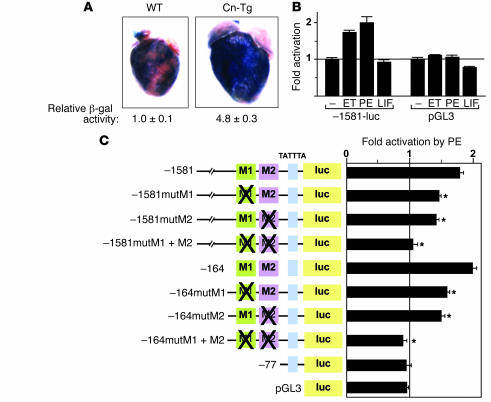
To evaluate whether the –1,581-bp STARS promoter was responsive to hypertrophic stress, we crossed Tg mice carrying the –1581STARS-lacZ transgene with Cn-Tg mice, which display marked cardiac hypertrophy (34). As shown in Figure 3A, the STARS-lacZ transgene was expressed much more strongly in mice of the Cn-Tg background than in wild-type littermates. The –1,581-bp upstream region of STARS also mediated inducible expression in response to endothelin-1 (ET) and phenylephrine (PE) in cultured cardiomyocytes (Figure 3B); mutation in either M1 or M2 reduced the response to PE of both the –1,581- and –164-bp promoters, and double M1 and M2 mutations abolished the response of both promoters to PE (Figure 3C). The activation of the –1,581-bp STARS promoter by ET or PE was comparable to that of the B-type natriuretic peptide (BNP) promoter (data not shown). We conclude that the M1 and M2 regions mediate both cardiac-specific and stress-inducible activation of STARS transcription.
Figure 3. The proximal promoter region mediates inducible expression of the STARS gene.
(A) LacZ staining and activity (n = 4) of adult Tg mouse hearts carrying construct 1 in wild-type or Cn-Tg mice. Original magnification, ×2. (B) Luciferase activity of cardiomyocytes transfected with –1581STARS-Luc or control pGL3 treated with 100 nM ET, 100 μM PE, or 1 nM leukemia inhibitory factor (LIF) for 24 hours. (C) Luciferase activity of cardiomyocytes transfected with –1581- or –164STARS-luc with or without mutations in the M1 or M2 regions and treated with 100 μM PE for 24 hours. *P < 0.05 versus the reporter without mutation.
MEF2C controls STARS expression.
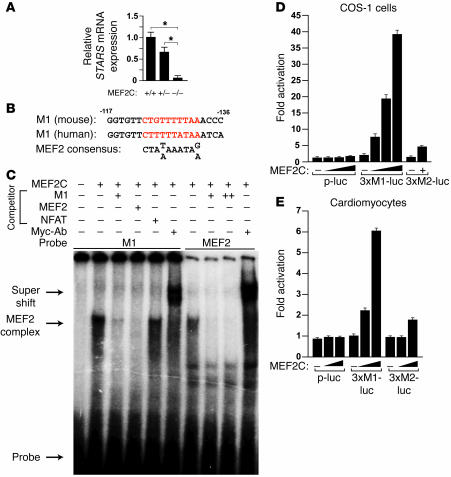
In microarray studies comparing gene expression profiles of hearts from wild-type and Mef2c-null embryos at E8.5, we found that STARS transcript levels were dramatically reduced in Mef2c mutants (data not shown). These findings were confirmed by real-time PCR (Figure 4A). Although the M1 and M2 regions do not contain typical conserved sequences for binding of MEF2 or other known myogenic transcription factors, the M1 region contains a sequence (TTAAAAACAG) resembling the MEF2-binding consensus site [CTA(T/A)AAATAG/A] (Figure 4B). In gel mobility shift assays, the M1 sequence bound MEF2C translated in vitro (Figure 4C), and an antibody against the MEF2 Myc-epitope tag caused a supershift (Figure 4C). The shifted band was completely eliminated by the unlabeled M1 sequence or the MEF2 consensus sequence from the desmin gene, but not by an unrelated NFAT-binding consensus sequence. A similar pattern of shifted bands was observed with labeled probes for the desmin MEF2-binding site (Figure 4C). MEF2C failed to bind the mutated M1 sequence or the M2 sequence (data not shown). Consistent with these results, MEF2C significantly increased expression of a luciferase reporter linked to 3 tandem M1 sites (3×M1-luc) fused to a minimum TATA box in both COS-1 cells and cardiomyocytes, whereas a reporter with 3 tandem M2 sites (3×M2-luc) or the minimum TATA alone (p-luc) failed to respond to MEF2C (Figure 4, D and E).
Figure 4. Regulation of the STARS promoter by MEF2.
(A) STARS mRNA expression in wild-type, Mef2c+/–, and Mef2c–/– embryos at E8.5 (n = 3). (B) Sequences of mouse and human M1 region aligned with MEF2 consensus binding site. Sequences are on the opposite strand relative to those in Figure 2F. (C) Gel mobility shift assay using in vitro translated myc-tagged MEF2C with radiolabeled STARS M1 DNA sequence or desmin MEF2-binding site as probe. Nonlabeled M1, desmin MEF2, or an unrelated NFAT consensus site was used as competitor (+ denotes 100-fold excess; ++ denotes 500-fold excess). Anti-myc antibody (Myc-Ab) was used to supershift the MEF2 complex. (D and E) Luciferase activity in COS-1 cells (D) and cardiomyocytes (E) cotransfected with luciferase reporters linked to multimerized M1 region (3×M1-luc), multimerized M2 region (3×M2-luc), or minimum TATA alone (p-luc) and MEF2C expression vector.
Expression of MEF2C increased the activity of –1581STARS-Luc and –164STARS-Luc by 40- and 14-fold, respectively, in COS-1 cells (Figure 5A). Mutation of the M1 sequence significantly reduced the responsiveness of both promoters to MEF2C, indicating that MEF2C activates the STARS promoter via the M1 region. Interestingly, a mutation in the M2 region, to which MEF2C cannot directly bind, also significantly reduced MEF2C responsiveness of the promoter, and mutation of both M1 and M2 regions further reduced MEF2C-inducible expression (Figure 5A). These results suggest that the M1 and M2 regions synergistically mediate MEF2C activation of STARS transcription. MEF2C significantly increased the activation of M1 and M2 tandemly fused to a luciferase reporter in both cardiac (data not shown) and noncardiac (COS-1 and NIH 3T3) cells, and a mutation in the M2 region reduced the activity of MEF2C, suggesting that the M2 region enhances MEF2 activity (Figure 5, B–D).
Figure 5. M1 and M2 region synergistically activate the STARS promoter.
(A) COS-1 cells cotransfected with –1581- or –164STARS-luc with or without mutations in the M1 or M2 region and an expression vector for MEF2C. (B) A schematic representation of tandem M1 plus M2 (M1+M2-luc) or M1 plus mutated M2 (M1+mutM2-luc) fused to a luciferase reporter. (C and D) Luciferase activity of COS-1 or NIH 3T3 cells, respectively, cotransfected with M1+M2-luc or M1+mutM2-luc and a MEF2C expression vector.
STARS induces ANP gene expression via SRF.
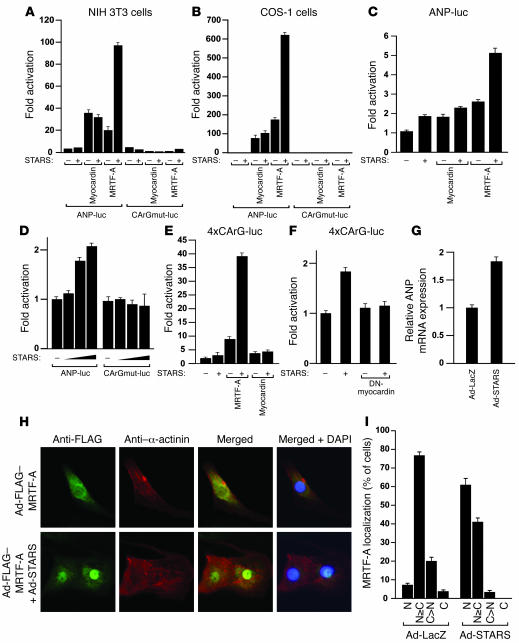
As STARS expression was increased during cardiac hypertrophy and cardiomyopathy, consistent with induction of SRF target genes, we examined whether STARS might activate the ANP promoter, which contains 2 SRF-binding sites (12, 35). In noncardiac cells, coexpression of STARS and the SRF coactivator MRTF-A significantly activated the ANP promoter (Figure 6, A and B). As reported previously (32, 33), STARS failed to enhance myocardin-mediated transcription of ANP (Figure 6, A and B) due to the constitutive localization of myocardin to the nucleus, which renders it resistant to nuclear translocation by STARS. Overexpression of STARS and MRTF-A in neonatal cardiomyocytes also synergistically increased ANP promoter activity (Figure 6C). Mutation of the 2 CArG boxes in the ANP promoter completely abolished the effect of STARS and MRTF-A (Figure 6, A, B, and D). STARS and MRTF-A also synergistically activated a reporter gene controlled by tandem CArG boxes (4×SRE-luciferase) (Figure 6E), and activation of 4×CArG-luciferase by STARS alone was completely inhibited by a dominant-negative mutant of myocardin that can inhibit the activity of both myocardin and MRTFs (Figure 6F), suggesting that the effect of STARS on SRF activity is mediated by endogenous MRTFs in cardiomyocytes.
Figure 6. STARS induces ANP expression by activation of SRF.
(A and B) Luciferase activity in NIH 3T3 (A) and COS-1 (B) cells cotransfected with ANP-luc or 2 CArG boxes mutated in ANP-luc (CArGmut-luc) and expression vectors encoding STARS, MRTF-A, and myocardin. (C–F) Luciferase activity of cardiomyocytes cotransfected with ANP-luc and expression vectors encoding STARS and MRTF-A (C); ANP-luc or CArGmut-luc and increasing amounts of STARS expression vector (D); multimerized CArG boxes linked to luciferase gene (4×CArG-luc) and STARS, MRTF-A, or myocardin expression vector (E); 4×CArG-luc and STARS and/or dominant-negative mutant of myocardin (DN-myocardin) expression vectors (F); or ANP mRNA expression in cardiomyocytes infected with recombinant adenovirus expressing STARS (Ad-STARS) or control lacZ (Ad-LacZ) (G), 48 hours after infection. Bars show mean ± SEM. (H) Immunostaining of cardiomyocytes infected with recombinant adenovirus expressing FLAG-tagged MRTF-A (Ad-FLAG-MRTF-A) with or without adenovirus expressing STARS (Ad-STARS), 48 hours after infection using anti-FLAG antibody (green) and anti–α-actinin monoclonal antibody (red). Original magnification, ×400. (I) Subcellular localization of MRTF-A in cardiomyocytes infected with adenovirus expressing STARS (Ad-STARS) or β-galactosidase (Ad-LacZ). C, cytoplasmic; C>N, cells with greater cytoplastmic than nuclear MRTF-A; N, nuclear; N≥C, cells with greater nuclear than cytoplasmic distribution of MRTF-A.
To examine the effect of STARS on endogenous ANP expression, we infected neonatal cardiomyocytes with recombinant adenovirus expressing STARS and analyzed ANP mRNA. Ectopic expression of STARS significantly increased endogenous ANP expression detected by real-time RT-PCR (Figure 6G). To confirm that STARS can induce nuclear translocation of MRTF-A in cardiomyocytes, we infected cardiomyocytes with recombinant adenovirus expressing MRTF-A with or without recombinant adenovirus expressing STARS. In the absence of serum, MRTF-A was located in both the cytoplasm and nucleus with some variability depending on the individual cell, and overexpression of STARS in cardiomyocytes enhanced the accumulation of MRTF-A in the nucleus (Figure 6, H and I).
STARS enhances the cardiac stress response.
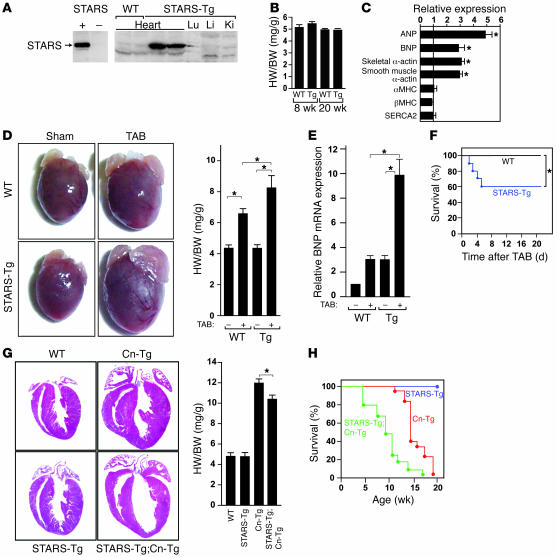
To examine the consequences of increased expression of STARS in vivo, we generated Tg mice using the α-MHC promoter to drive cardiac expression of STARS. We obtained 2 independent Tg lines that displayed similar phenotypes, and we describe 1 of the lines here. Hearts isolated from α-MHC-STARS–Tg mice showed an approximately 20-fold increase in STARS mRNA expression in the heart compared with non-Tg littermates (Figure 7A). Heart weight/body weight ratios and total body weight of α-MHC-STARS–Tg mice and wild-type littermates were not significantly different (Figure 7B). Hearts of α-MHC-STARS–Tg mice also did not show histological abnormalities (data not shown). Nevertheless, several fetal cardiac genes known to be direct targets of SRF were upregulated in α-MHC-STARS–Tg mice (Figure 7C).
Figure 7. Heightened sensitivity of STARS-Tg mice to hypertrophic stimuli.
(A) Western blot of STARS protein expression in tissues of α–MHC-STARS–Tg mice. Left panel shows antibody control of 293T cells transfected with or without a STARS expression vector. Lu, lung; Li, liver; Ki, kidney. (B) Heart weight/body weight (HW/BW) ratios of α–MHC-STARS–Tg mice and control wild-type littermates at 8 (n = 12) and 20 (n = 4) weeks of age. (C) Cardiac gene expression determined by RT-PCR using total RNA extracted from α–MHC-STARS–Tg mice and control wild-type littermate at 8 weeks of age (n = 6 each). *P < 0.05 versus control wild-type littermates. (D) Hearts and HW/BW ratio of α–MHC-STARS–Tg mice and wild-type littermates at 10 weeks of age with or without TAB. n = 8 (Tg) and n = 6 (WT). Original magnification, ×2. (E) BNP mRNA expression in the hearts of α–MHC-STARS–Tg mice and wild-type littermates with or without TAB (n = 3). (F) Kaplan-Meier survival curve of α–MHC-STARS–Tg and WT mice after TAB. Mice that died within 24 hours after TAB were excluded. (G) Hearts (H&E-stained) and HW/BW ratios of α–MHC-STARS–Tg mice, Cn-Tg mice, α–MHC-STARS–Tg;Cn-Tg mice, and WT mice (n = 6 in each group). Original magnification, ×2. (H) Kaplan-Meier survival curve of α–MHC-STARS–Tg mice, Cn-Tg mice, and α–MHC-STARS–Tg;Cn-Tg mice. *P < 0.05.
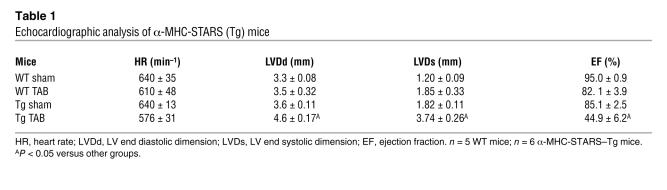
To determine whether increased expression of STARS in hypertrophic hearts is beneficial or deleterious, we subjected α-MHC-STARS–Tg mice to pressure overload by TAB and observed an exaggerated hypertrophic response (Figure 7D). In addition, BNP expression was augmented in α-MHC-STARS–Tg mice subjected to TAB (Figure 7E), and echocardiography showed that cardiac systolic function was markedly decreased compared with that in wild-type mice subjected to TAB, as determined by the decrease in ejection fraction from 82.1% ± 3.9% to 44.9% ± 6.2% (Table 1 and Supplemental Methods; supplemental material available online with this article; doi:10.1172/JCI31240DS1). Measurements of the diastolic LV diameter showed an increase from 3.5 ± 0.32 mm to 4.6 ± 0.17 mm, indicating significant LV chamber dilation in α-MHC-STARS–Tg mice subjected to TAB compared with non-Tg mice with TAB (Table 1 and Supplemental Methods). Following TAB, α-MHC-STARS–Tg mice also displayed increased mortality compared with non-Tg mice (Figure 7F).
Table 1 .
Echocardiographic analysis of α-MHC-STARS (Tg) mice
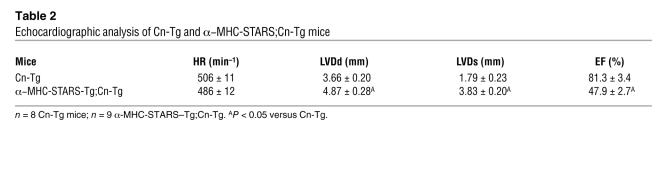
We also crossed α-MHC-STARS–Tg mice to Cn-Tg mice. In the Cn-Tg background, α-MHC-STARS–Tg mice showed a slightly decreased heart weight/body weight ratio compared with Cn-Tg mice (Figure 7G). The double-Tg hearts showed thinner ventricular walls with greater ventricular dilatation than the hearts from Cn-Tg mice (Figure 7G). Echocardiography of Cn-Tg;α-MHC-STARS–Tg mice at 8 weeks of age showed a reduction in wall thickness, an increase in diastolic and systolic ventricular diameters, and a decrease in systolic function compared with Cn-Tg mice, which showed marked wall hypertrophy with preserved systolic function (Table 2 and Supplemental Methods). Furthermore, Cn-Tg;α-MHC-STARS–Tg mice showed decreased survival relative to Cn-Tg mice (Figure 7H). We conclude that increased expression of STARS enhances the sensitivity of the heart to stress signals.
Table 2 .
Echocardiographic analysis of Cn-Tg and α–MHC-STARS;Cn-Tg mice
Discussion
The results of this study demonstrate that STARS, which encodes a striated muscle–restricted cytoskeletal protein, serves as a stress-inducible target gene of MEF2. Increased expression of STARS in cardiomyocytes activated the expression of SRF-dependent fetal cardiac genes and led to an accelerated deterioration of cardiac function under hypertrophic conditions. These findings suggest that STARS modulates the cardiac stress response and couples MEF2 and SRF signaling.
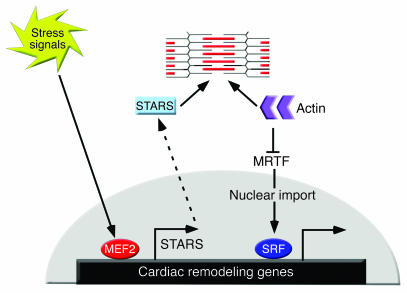
STARS stimulates SRF activity, at least in part, by depleting the G-actin pool, thereby releasing MRTFs to translocate to the nucleus, where they associate with SRF and stimulate SRF target genes (33). In addition to regulating stress-responsive genes such as ANP, SRF regulates the expression of the sarcomeric actin genes, which contributes to further assembly of sarcomeres, a hallmark of cardiac hypertrophy. Thus, the upregulation of STARS by MEF2 in response to stress signals would be expected to promote actin polymerization, thereby further stimulating SRF activity and actin expression. A model to account for such a feed-forward mechanism of signaling between the contractile apparatus and the hypertrophic gene program is shown in Figure 8.
Figure 8. A model for the role of STARS as a cytoskeletal intermediary between MEF2 and SRF.
MEF2 regulates STARS expression and STARS stimulates SRF activity by sequestering actin monomers, thereby freeing MRTFs to translocate to the nucleus and promote SRF-dependent gene expression. Sustained increase in STARS expression results in the alteration of the cardiac gene program, which may facilitate the transition to cardiac dysfunction.
Regulation of STARS expression by MEF2.
Both basal and inducible expression of STARS are mediated, at least in part, by a MEF2-binding site in the STARS promoter. Supporting this finding, expression of STARS is diminished in Mef2c-null embryos. In addition to the essential MEF2 site (M1) in the STARS promoter, a second site (M2) cooperates with the M1 site but does not bind MEF2 directly, suggesting that an M2-binding factor cooperates with MEF2 to activate STARS transcription. This premise seems contradictory to the finding that a mutation in the M1 region does not completely abolish induction of STARS expression by MEF2. However, it should be noted that the proximal TATA box also resembles a MEF2 consensus sequence, and previous studies have shown that MEF2 can activate transcription through TATA sequences (36, 37). Thus, in the absence of M1, MEF2 may bind to this TATA box–like element and activate STARS transcription synergistically with a factor that binds to the M2 region. MEF2 might also act through both the M1 and TATA box to activate transcription of the STARS gene.
STARS enhances SRF activity in cardiomyocytes.
STARS promotes nuclear translocation of MRTF-A and synergizes with MRTF-A to stimulate SRF-mediated transcription (33). A dominant-negative myocardin mutant that blocks the activity of myocardin and MRTFs impedes STARS activation of SRF in cardiomyocytes, suggesting that increased expression of STARS in hypertrophic hearts contributes to the activation of SRF. Indeed, cardiac-specific overexpression of STARS increased the expression of SRF-dependent fetal cardiac genes in the absence of cardiac hypertrophy (Figure 7C). Previously, we showed that myocardin, which is expressed in cardiomyocytes, is involved in activation of the fetal gene program during cardiac hypertrophy (38). Given that myocardin and MRTFs can form heterodimeric complexes to activate SRF (21), we postulate that the STARS/MRTF pathway and other pathways activating myocardin may cooperate to enhance SRF activity during cardiac remodeling.
STARS sensitizes the heart to stress.
Our results show that elevated expression of STARS in the hearts of α-MHC-STARS–Tg mice resulted in heightened sensitivity to cardiac stress. Increased expression of STARS in response to hypertrophic stimuli may initially serve as a compensatory response to increased actin content to maintain cytoskeletal integrity and sustain systolic function against increased afterload, but excessive expression of STARS leads to cardiac maladaptation. In this regard, cardiac-specific overexpression of SRF causes severe dilated cardiomyopathy accompanied by activation of the fetal gene program (16), supporting our finding that sustained activation of SRF is deleterious to cardiac function. In contrast, cardiac-specific deletion of SRF in the embryonic heart results in cardiac defects (14, 15). Deletion of SRF from the adult heart also causes a dilated cardiomyopathic phenotype (39), indicating that SRF is necessary for normal cardiac development and maintenance of normal function in the postnatal heart. Thus, both loss and gain of function of SRF are deleterious to cardiac structure and function.
Collectively, the results of this study demonstrate that STARS is a stress-inducible target of MEF2 and activator of SRF. Sustained overexpression of STARS facilitates functional deterioration in hypertrophic hearts, implicating STARS in the transition from cardiac hypertrophy to heart failure. Further investigation of the role of STARS will provide insights into the mechanisms whereby MEF2 and SRF regulate cardiac function during development and disease, as well as the signaling networks that couple the sarcomere with the cardiac genome.
Methods
Plasmid constructs.
DNA fragments from various portions of the 5' upstream region of the mouse STARS gene relative to the ATG codon (–1,581 to +53 bp, –1,043 to +53 bp, and –164 to +53 bp) were isolated by PCR using the mouse genome as template and ligated upstream of a lacZ gene or the luciferase reporter pGL3 (Promega). Primer sequences and plasmid construct designs are described in Supplemental Methods.
PCR-based mutagenesis in M1 and M2 sequences of the STARS promoter was performed according to standard procedures using primers described in Supplemental Methods. The –452-bp human ANP promoter–luciferase reporter gene was previously reported (40). PCR-based mutagenesis of the 2 CArG sequences of the ANP promoter was performed using primers described in Supplemental Methods. The pcDNA3-based expression vectors for mouse STARS and MRTF-A were previously reported (32, 33).
Cell culture, luciferase assay, adenovirus infection, and immunocytochemistry.
Primary neonatal rat ventricular myocytes were isolated and grown as described previously (34). Twenty-four hours after plating, the myocytes were transfected with 200 ng of reporter plasmid and 200 ng of expression vectors for 12 hours using GeneJammer (Stratagene), unless indicated otherwise. After transfection, serum was removed from the growth medium. Six hours after serum deprivation, ET-1 (100 nM), PE (100 μM), or vehicle was added, and the cells were maintained for an additional 48 hours.
Recombinant adenovirus was generated using cDNAs encoding mouse STARS; or MRTF-A were cloned into pAC-CMV vector and the resultant constructs were cotransfected into HE 293 cells with pJM17 using FuGENE 6 (Roche Diagnostics). Clonal populations of adenoviruses were amplified by reinfecting HEK 293 cells, and the viral preparations were titered using the agar overlay method.
Cardiomyocytes grown on coverslips in 6-well dishes were infected 36 hours after plating with recombinant adenovirus at a multiplicity of infection of 5 for 6 hours and subsequently maintained in serum-containing medium for 36 hours. After the medium was changed to serum-free medium, cells were further incubated for 12 hours and then fixed with 4% formaldehyde in PBS. Immunocytochemistry was performed as previously described (32, 33), and detailed protocols are presented in Supplemental Methods. Luciferase assays were performed using the Luciferase Assay System (Promega) and FluoReporter lacZ/Galactosidase Quantitation Kit (Invitrogen) according to the manufacturers’ instructions.
EMSA.
EMSAs were performed as previously described (24) with double-stranded oligonucleotides and method modifications described in Supplemental Methods.
Tg mice, TAB, echocardiography, and histological analysis.
Tg mice harboring various DNA fragments of the STARS gene regulatory region fused to the minimum HSP68 basal promoter and the lacZ reporter gene were generated using standard procedures (41, 42). TAB of 6- to 8-week-old male mice was performed as previously described (43). Histological analysis was performed as described previously (6). Echocardiography was performed using a Hewlett-Packard Sonos 5500 Ultrasound system as described previously (6). All animal protocols used in this study were approved by the University of Texas Southwestern Institutional Animal Care and Use Committee. Further procedural details are available in Supplemental Methods.
Real time RT-PCR.
Total RNA was isolated from cultured neonatal ventricular myocytes or mouse hearts, using TRI zol, following the manufacturer’s protocol. Real-time One-Step RT-PCR was performed with 20–100 ng total RNA using TaqMan One-Step RT-PCR Master Mix reagent and primers (Applied Biosystems). Sequences of primers and probes are shown in Supplemental Methods.
Human DNA samples.
Tissue samples of LVs from hearts of unidentified individuals diagnosed as having idiopathic cardiomyopathy (3 males; average age 57 years) or nonfailing hearts (2 males, 1 female; average age 45 years) were obtained from Gilead. This study involved the use of existing pathological specimens, and the information was recorded in such a manner that the subjects cannot be identified directly or through identifiers linked to the subjects. This study qualified for exempt review by the Institutional Review Board of the University of Texas Southwestern Medical Center in accordance with Department of Health and Human Services regulations.
Statistics.
Data are presented as mean ± SEM. An unpaired 2-tailed Student’s t test was used for comparison between 2 groups, and ANOVA with post-hoc Fisher’s test was used for comparison among groups. P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Yongli Kong for assistance with TAB; Robert Gerard for providing STARS recombinant adenovirus; Esther Creemers, Tomasa Barrientos, Jeffrey Spencer, and Akiko Arai for scientific advice and plasmid constructs; Guangrong Lu for technical assistance; Angela Diehl for graphics; and Jennifer Brown for editorial assistance. This work was supported by grants from the NIH, the Donald W. Reynolds Cardiovascular Clinical Research Center, the American Heart Association, and the Robert A. Welch Foundation to E.N. Olson. G.C. Teg Pipes was supported by an NIH postdoctoral fellowship. K. Kuwahara was supported by a research fellowship from the Uehara Memorial Foundation.
Footnotes
Nonstandard abbreviations used: ANP, atrial natriuretic peptide; BNP, B-type natriuretic peptide; ET, endothelin-1; MEF2, myocyte enhancer factor–2; α-MHC, α–myosin heavy chain; MRTF, myocardin-related transcription factor; NFAT, nuclear factor of activated T cells; PE, phenylephrine; SRF, serum response factor; STARS, striated muscle activator of Rho signaling; TAB, thoracic aortic banding.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:1324–1334 (2007). doi:10.1172/JCI31240
References
- 1.Chien K.R., Olson E.N. Converging pathways and principles in heart development and disease: CV@CSH. Cell. 2002;110:153–162. doi: 10.1016/s0092-8674(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 2.Frey N., Olson E.N. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 3.Seidman J.G., Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 4.Granzier H., Wu Y., Siegfried L., LeWinter M. Titin: physiological function and role in cardiomyopathy and failure. Heart Fail. Rev. 2005;10:211–223. doi: 10.1007/s10741-005-5251-7. [DOI] [PubMed] [Google Scholar]
- 5.Knoll R., et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 6.Frey N., et al. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 2004;10:1336–1343. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 7.Purcell N.H., et al. Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. Mol. Cell. Biol. 2004;24:1081–1095. doi: 10.1128/MCB.24.3.1081-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippar U., et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Lange S., et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 10.Pipes G.C., Creemers E.E., Olson E.N. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 11.Miano J.M. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 12.Sprenkle A.B., Murray S.F., Glembotski C.C. Involvement of multiple cis elements in basal- and alpha-adrenergic agonist-inducible atrial natriuretic factor transcription. Roles for serum response elements and an SP-1-like element. Circ. Res. 1995;77:1060–1069. doi: 10.1161/01.res.77.6.1060. [DOI] [PubMed] [Google Scholar]
- 13.Nelson T.J., Balza R., Jr., Xiao Q., Misra R.P. SRF-dependent gene expression in isolated cardiomyocytes: regulation of genes involved in cardiac hypertrophy. J. Mol. Cell. Cardiol. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Parlakian A., et al. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol. Cell. Biol. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miano J.M., et al. Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., et al. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H1782–H1792. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 17.Chang J., et al. Inhibitory cardiac transcription factor, SRF-N, is generated by caspase 3 cleavage in human heart failure and attenuated by ventricular unloading. Circulation. . 2003;108:407–413. doi: 10.1161/01.CIR.0000084502.02147.83. [DOI] [PubMed] [Google Scholar]
- 18.Wang D., et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang D.Z., et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cen B., et al. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Wang D.Z., Pipes G.C., Olson E.N. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S., Wang D.Z., Wang Z., Richardson J.A., Olson E.N. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miralles F., Posern G., Zaromytidou A.I., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 24.Gossett L.A., Kelvin D.J., Sternberg E.A., Olson E.N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black B.L., Olson E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 26.McKinsey T.A., Zhang C.L., Olson E.N. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 27.Lin Q., Schwarz J., Bucana C., Olson E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naya F.J., et al. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 29.Wu H., et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passier R., et al. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J., et al. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J. Biol. Chem. 2006;281:9152–9162. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 32.Arai A., Spencer J.A., Olson E.N. STARS, a striated muscle activator of Rho signaling and serum response factor-dependent transcription. J. Biol. Chem. 2002;277:24453–24459. doi: 10.1074/jbc.M202216200. [DOI] [PubMed] [Google Scholar]
- 33.Kuwahara K., Barrientos T., Pipes G.C., Li S., Olson E.N. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol. Cell. Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molkentin J.D., et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin S., Paradis P., Aries A., Nemer M. Serum response factor-GATA ternary complex required for nuclear signaling by a G-protein-coupled receptor. Mol. Cell. Biol. 2001;21:1036–1044. doi: 10.1128/MCB.21.4.1036-1044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grayson J., Williams R.S., Yu Y.T., Bassel-Duby R. Synergistic interactions between heterologous upstream activation elements and specific TATA sequences in a muscle-specific promoter. Mol. Cell. Biol. 1995;15:1870–1878. doi: 10.1128/mcb.15.4.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leibham E., et al. Binding of TFIID and MEF2 to the TATA element activates transcription of the Xenopus MyoDa promoter. Mol. Cell. Biol. 1994;14:686–699. doi: 10.1128/mcb.14.1.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing W., et al. Myocardin induces cardiomyocyte hypertrophy. Circ. Res. 2006;98:1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
- 39.Parlakian A., et al. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 2005;112:2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778. [DOI] [PubMed] [Google Scholar]
- 40.Kuwahara K., et al. The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol. Cell. Biol. 2001;21:2085–2097. doi: 10.1128/MCB.21.6.2085-2097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa O., et al. Centronuclear myopathy in mice lacking a novel muscle-specific protein kinase transcriptionally regulated by MEF2. Genes Dev. 2005;19:2066–2077. doi: 10.1101/gad.1338705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng T.C., Hanley T.A., Mudd J., Merlie J.P., Olson E.N. Mapping of myogenin transcription during embryogenesis using transgenes linked to the myogenin control region. J. Cell Biol. 1992;119:1649–1656. doi: 10.1083/jcb.119.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill J.A., et al. Cardiac hypertrophy is not a required compensatory response to short-term pressure overload. Circulation. 2000;101:2863–2869. doi: 10.1161/01.cir.101.24.2863. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.