Abstract
Erythropoiesis is critically dependent on erythropoietin (EPO), a glycoprotein hormone that is regulated by hypoxia-inducible factor (HIF). Hepatocytes are the primary source of extrarenal EPO in the adult and express HIF-1 and HIF-2, whose roles in the hypoxic induction of EPO remain controversial. In order to define the role of HIF-1 and HIF-2 in the regulation of hepatic EPO expression, we have generated mice with conditional inactivation of Hif-1α and/or Hif-2α (Epas1) in hepatocytes. We have previously shown that inactivation of the von Hippel–Lindau tumor suppressor pVHL, which targets both HIFs for proteasomal degradation, results in increased hepatic Epo production and polycythemia independent of Hif-1α. Here we show that conditional inactivation of Hif-2α in pVHL-deficient mice suppressed hepatic Epo and the development of polycythemia. Furthermore, we found that physiological Epo expression in infant livers required Hif-2α but not Hif-1α and that the hypoxic induction of liver Epo in anemic adults was Hif-2α dependent. Since other Hif target genes such phosphoglycerate kinase 1 (Pgk) were Hif-1α dependent, we provide genetic evidence that HIF-1 and HIF-2 have distinct roles in the regulation of hypoxia-inducible genes and that EPO is preferentially regulated by HIF-2 in the liver.
Introduction
The glycoprotein erythropoietin (EPO) is essential for the regulation of red blood cell mass in response to changes in tissue oxygenation. EPO induces erythropoiesis through the stimulation of erythroid precursor cell viability, proliferation, and differentiation, thus enhancing the oxygen-carrying capacity of blood (review in ref. 1). Lack of Epo during murine development results in embryonic lethality at E13.5 as a result of cardiac failure and anemia (2, 3). Clinically, dysregulated EPO expression results in the development of anemia when serum EPO levels are inadequately low or polycythemia as a result of EPO overproduction.
EPO expression is tightly regulated by developmental, tissue-specific, and physiological cues (1). During development, the physiological source of EPO switches from the fetal liver to the kidney in adults, the timing of which is species dependent (4–7). Although the kidney is the primary physiologic source of EPO in the adult, the liver maintains the capacity to express EPO. Hepatocytes are the primary cell type responsible for Epo expression in the liver (8). During normal erythropoiesis, the ratio between kidney and liver Epo levels in the adult is 9:1 (9). However, under conditions of severe hypoxia, it has been estimated that liver Epo production may account for more than 33% of total Epo (10).
The primary physiological stimulus of increased EPO gene transcription is tissue hypoxia, which can induce up to a 1,000-fold increase in circulating serum EPO levels (1). Studies aimed at isolating factors responsible for the hypoxia-mediated induction of EPO identified the heterodimeric basic helix-loop-helix (bHLH) transcription factor hypoxia-inducible factor–1 (HIF-1) (11–13). HIF-1 belongs to the PAS (PER/ARNT/SIM [PER/aryl-hydrocarbon-receptor nuclear translocator/single minded]) family of hypoxia-regulated transcription factors and consists of an oxygen-sensitive α subunit and the constitutively expressed β subunit ARNT (14). Together with HIF-2α (also known as EPAS-1, HLF), HIF-1α facilitates oxygen delivery and cellular adaptation to hypoxia by stimulating erythropoiesis, angiogenesis, and anaerobic glucose metabolism (15, 16). Under normoxia, HIF-1α and HIF-2α are targeted for proteasomal degradation by the von Hippel–Lindau (VHL) tumor suppressor pVHL, which functions as the substrate recognition component of an E3 ubiquitin-ligase complex. pVHL-mediated targeting of HIF-α requires oxygen- and iron-dependent hydroxylation of specific proline residues within the oxygen-dependent degradation domain (ODD) of HIF-α by specific prolyl-4-hydroxylase domain–containing (PHD-containing) proteins (reviewed in ref. 17). In the absence of either oxygen or HIF ubiquitination (i.e., as a result of mutated pVHL), HIF-α subunits are stabilized and translocate to the nucleus, where they heterodimerize with ARNT and activate transcription upon binding to hypoxia response elements (HREs) located within regulatory elements of HIF target genes (reviewed in refs. 15, 16).
The role of HIF-1 and HIF-2 in the hypoxic induction of hepatic EPO remains controversial. Although HIF-1 was initially purified from human hepatoma (Hep3B) cells as the HIF that bound to an 18-nucleotide fragment of the 3' EPO regulatory element containing the HRE, which appears to specifically control hepatic EPO expression (18), the hypoxic induction of endogenous EPO in the same cell line was recently found to be largely dependent on HIF-2 and not HIF-1 using an siRNA knockdown approach (13, 19). While the regulation of EPO in Hep3B cells is dependent on HIF-2, EPO induction in HepG2 hepatoma and SH-SY5Y neuroblastoma cells was suggested to be HIF-1 dependent, indicating cell type–specific differences (M.P. Biju and V.H. Haase, unpublished observations; ref. 20). It has been proposed that in vivo HIF-1 plays a role in the physiologic regulation of Epo expression in the embryo and in the hypoxic regulation of Epo in the retina and liver (21–23). In contrast, we have previously shown that hepatic overproduction of Epo in a mouse model of VHL-associated polycythemia is not dependent on HIF-1 but requires ARNT, suggesting a dominant role for HIF-2 (24). In order to define the role of HIF-1 and HIF-2 in hepatic EPO regulation, we utilized Cre-loxP–mediated recombination to inactivate murine HIF-1α or HIF-2α (hereafter referred to as Hif-1α and Hif-2α) in the livers of mice deficient in VHL syndrome homolog (Vhlh, the murine ortholog of the human VHL gene) and WT mice.
In this report, we demonstrate by direct genetic comparison that inactivation of Hif-2α, but not Hif-1α, is sufficient to suppress hepatic Epo induction in both Vhlh deficient and WT adult mice that were made acutely anemic by phenylhydrazine (PHZ) treatment and phlebotomy. Additionally, we show that the absence of hepatic Hif-2α, in contrast to Hif-1α, resulted in a suppression of physiologic baseline Epo levels in the infant liver. Furthermore, we propose that HIF-2 dominance is the result of preferential HIF-2 binding to the HRE located within the endogenous EPO 3' enhancer element. Our results establish a principal role for HIF-2 in the hypoxic regulation of liver EPO in vivo and have implications for therapeutic strategies in the treatment of anemia associated with chronic renal disease aimed at pharmacological manipulations of the HIF oxygen-sensing pathway.
Results
Efficient recombination of the Hif-1α and Hif-2α conditional alleles in hepatocytes.
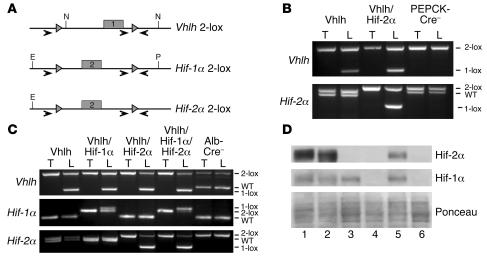
Hepatocytes are the primary source of extrarenal EPO in response to hypoxia and anemia. They express 2 HIF-α homologs (HIF-1α and HIF-2α) that have been previously characterized as hypoxia-responsive transcriptional activators, which have the capacity to activate EPO expression (25–27). In order to determine the requirement for HIF-1α and HIF-2α in the hypoxic induction of hepatic EPO expression in vivo, we conditionally inactivated Hif-1α and Hif-2α in hepatocytes of Vhlh-deficient and WT mice. Conditional alleles for both Hif-1α and Hif-2α (Hif-1α 2-lox, Hif-2α 2-lox) contain LoxP sites that flank exon 2, which encodes the bHLH DNA binding domain (Figure 1A), resulting in an out-of-frame deletion of exon 2 and inactivation of Hif-1α and Hif-2α, respectively, upon Cre-mediated recombination (28, 29).
Figure 1. Generation of mice deficient for Vhlh, Vhlh/Hif-1α, Vhlh/Hif-2α, and Vhlh/ Hif-1α/Hif-2α in the liver.
(A) Genomic maps of the Vhlh (2-lox), Hif-1α (2-lox), and Hif-2α (2-lox) conditional alleles. Numbered boxes represent exons targeted for deletion. The locations of LoxP sites and primers used to amplify the conditional allele (2-lox), recombined allele (1-lox), and WT allele are shown with gray triangles and arrows, respectively. N, NcoI; E, EcoR1; P, PstI. (B) Genomic PCR analysis of DNA isolated from the tail (T) and liver (L) of PEPCK-Cre mutant mice. (C) Genomic PCR analysis of DNA isolated from the tail and liver of 4-week-old albumin-Cre mutant mice. Note that mice were bred in a mixed genetic background. Occasionally 2 different Hif-2α WT alleles were detected by genomic PCR analysis, suggesting a polymorphism in the amplified region. (D) Western blot analysis of nuclear protein isolated from livers of the following albumin-Cre mutant littermates: lane 1, -Vhlh haploinsufficient for Hif-1α; lane 2, -Vhlh/Hif-1α haploinsufficient for Hif-2α; lane 3, -Vhlh/Hif-2α; lane 4, -Vhlh/Hif-1α/Hif-2α; lane 5, -Vhlh/Arnt; and lane 6, Cre-negative mice. Note that the recombination efficiency of the Hif-1α conditional allele is best determined by comparing Cre-negative control mice to Vhlh/Hif-1α/Hif-2α mutant mice due to the presence of nonrecombined inflammatory cells in albumin-Vhlh/Hif-1α mutant mice.
For the hepatocyte-specific inactivation of Hif-1α, Hif-2α, and Vhlh, we used PEPCK-Cre and albumin-Cre transgenic lines. PEPCK-Cre transgenic mice express Cre-recombinase under control of a mutated phosphoenolpyruvate carboxykinase promoter, which is expressed in periportal hepatocytes targeting approximately 20% of hepatocytes (24). As previously shown, PEPCK-Cre–mediated inactivation of Vhlh results in increased hepatic Epo and development of polycythemia that was found to be Hif-1α independent (24). To determine the requirement for Hif-2α in hepatic Epo expression and the development of polycythemia in PEPCK-Vhlh mutant mice, we generated PEPCK-Vhlh/Hif-2α mutant mice. Similar to the Vhlh conditional allele, the Hif-2α 2-lox allele was efficiently recombined in PEPCK-Vhlh/Hif-2α mutant livers as determined by genomic PCR analysis (Figure 1B).
Albumin-Cre transgenic mice express Cre-recombinase in the majority of hepatocytes (>80%) under the control of the albumin promoter (30). Recombination of the Vhlh 2-lox allele by albumin-Cre results in severe liver pathology and premature morbidity by 4–6 weeks of age (24). Albumin-Cre–mediated recombination of the Hif-1α and Hif-2α conditional alleles was readily detectable in adult albumin-Vhlh/Hif-1α, albumin-Vhlh/Hif-2α, and albumin-Vhlh/Hif-1α/Hif-2α mutant livers by genomic PCR analysis (Figure 1C) and resulted in complete loss of Hif-1α and Hif-2 α protein in albumin-Vhlh/Hif-1α/Hif-2α mutant livers (Figure 1D, compare lanes 1 and 3 with lane 4). Due to the high recombination efficiency, albumin–Hif-1α and –Hif-2α mutant mice were used to investigate the requirement of Hif-1α and Hif-2α in the induction of hepatic Epo and other Hif-regulated genes in anemic mice.
Inactivation of Hif-2α suppresses the development of polycythemia in Vhlh-deficient mice.
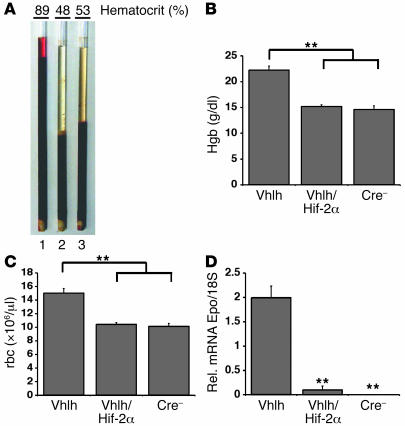
Patients with VHL disease are predisposed to the development of polycythemia that is associated with paraneoplastic EPO production from tumors including renal cell carcinomas, hemangioblastomas, and pheochromocytomas (31). We have recently reported that Vhlh gene deletion in the liver results in Hif activation (both Hif-1 and Hif-2) and the development of polycythemia associated with increased hepatic Epo expression (24, 32). Moreover, we observed that the development of polycythemia in Vhlh-deficient mice occurred in an Hif-1–independent and Arnt-dependent manner, suggesting a predominant role for Hif-2 (24). To determine the requirement for Hif-2 in the development of polycythemia in PEPCK-Vhlh mutant mice, we compared hematocrit values, hemoglobin concentrations, and red blood cell numbers in PEPCK-Vhlh, PEPCK-Vhlh/Hif-2α, and PEPCK-Cre–negative mice. We observed that PEPCK-Vhlh/Hif-2α and WT mice exhibited similar hematocrit values (51% ± 9.5% and 53% ± 6.5%, respectively; Figure 2A), hemoglobin concentrations (15.18 ± 0.3 g/dl and 14.6 ± 0.7 g/dl, respectively; Figure 2B), and red blood cell numbers (10.425 × 106 ± 0.2 × 106/μl and 10.133 × 106 ± 0.43 × 106/μl, respectively; Figure 2C). In contrast, PEPCK-Vhlh littermates developed erythrocytosis, with average hematocrit values of 78% ± 14.3% (Figure 2A), hemoglobin concentrations of 22.25 ± 0.8 g/dl (Figure 2B), and red blood cell numbers of 15.025 × 106 ± 0.7 × 106/μl (Figure 2C). From these data, we conclude that despite constitutive activation of both Hif factors, Hif-2α is required for the development of polycythemia in PEPCK-Vhlh mutant mice.
Figure 2. Inactivation of Hif-2α suppresses the development of polycythemia in PEPCK-Vhlh mutant mice.
(A) Photograph of centrifuged microcapillary tubes containing blood from representative PEPCK-Vhlh (hematocrit 89%; 1), PEPCK-Vhlh/Hif-2α (hematocrit 48%; 2), and control (hematocrit 53%; 3) mice. (B and C) Elevated hemoglobin (Hgb) and red blood cell numbers in PEPCK-Vhlh mutant mice were suppressed in PEPCK-Vhlh/Hif-2α mutant mice. Shown are hemoglobin concentrations and red blood cell (rbc) numbers in blood collected from PEPCK-Cre mutant and control (Cre–) mice determined by a complete blood count analyzer. Shown are the mean values for 6 individual mice. Error bars represent SEM. (D) Inactivation of Hif-2α significantly decreases Epo transcript levels in PEPCK-Vhlh mutant livers. Shown are relative Epo mRNA transcript levels normalized to 18S in the livers of PEPCK-Cre mutant and Cre-negative mice. Bars represent mean mRNA transcript levels of 3 mice per group. Error bars represent SEM. **P < 0.001 compared with PEPCK-Vhlh as determined by Student’s t test.
Because the development of polycythemia in PEPCK-Vhlh mutants is associated with increased Epo expression, we next determined whether inactivation of Hif-2α is sufficient to suppress Epo transcript levels. Real-time PCR analysis revealed that relative hepatic Epo mRNA levels in PEPCK-Vhlh/Hif-2α mutants were significantly reduced compared with those in PEPCK-Vhlh mutant mice (P < 0.001) (Figure 2D). Although Epo transcripts were detectable in PEPCK-Vhlh/Hif-2α mutant livers, they were not significantly different from Epo levels in control mice (P = 0.268). In conclusion, inactivation of Hif-2α is sufficient to suppress hepatic Epo expression in Vhlh-deficient livers despite increased Hif-1α activity, suggesting that Hif-2 is the dominant Hif with regard to hepatic Epo regulation.
Hif-1 and Hif-2 activate distinct target genes in Vhlh-deficient livers.
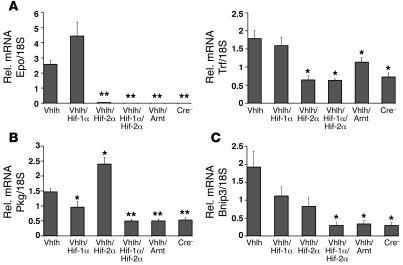
In order to determine whether other Hif target genes are preferentially regulated by Hif-2 in hepatocytes, we examined Hif target gene expression levels in the livers of albumin-Cre mutants by real-time PCR. Similar to what was observed in PEPCK-Vhlh mutant mice, Epo expression in albumin-Vhlh mutants was Hif-2α dependent (Figure 3A). Epo mRNA levels in albumin-Vhlh/Hif-1α/Hif-2α and albumin-Vhlh/Arnt mutant mice were not significantly different from those in albumin-Vhlh/Hif-2α or control mice, suggesting that the Hif-1α/Arnt heterodimer plays only a minor role in the transcriptional regulation of hepatic Epo (Figure 3A).
Figure 3. Differential HIF target gene expression in albumin-Vhlh/Hif-1α– and albumin-Vhlh/Hif-2α–deficient livers.
Relative mRNA levels for HIF target genes in the liver of albumin-Cre mutant mice determined by real-time PCR. (A) Inactivation of Hif-2α significantly decreases the expression of HIF target genes regulating erythropoiesis (Epo) and iron transport (Trf). (B) Inactivation of Hif-1α decreases the expression of the glycolytic target gene Pgk. (C) Inactivation of both Hif-1α and Hif-2α decreases the expression of the proapoptotic gene Bnip3. Bars represent the mean mRNA transcript level of 3 mice for the albumin-Vhlh/Hif-1α/Hif-2α group and 4 mice for all other groups. Error bars represent SEM. *P < 0.05, **P < 0.001 compared with albumin-Vhlh mutants as determined by Student’s t test.
Similar to that of Epo, expression of the HIF target gene transferrin (Trf), which encodes an iron transport protein, was induced in an Hif-2α–specific manner in albumin-Vhlh mutant livers (Figure 3A; refs. 22, 33). Because Hif-2α was required for the activation of both Epo and Trf expression, we next examined the regulation of 2 Hif target genes that have been previously characterized as preferential Hif-1α targets in vitro, the glycolytic enzyme (phosphoglycerate kinase) Pgk and the proapoptotic gene BCL2/adenovirus E1B–interacting protein 1, NIP3 (Bnip3) (34, 35). Inactivation of Hif-1α significantly decreased Pgk mRNA levels in albumin-Vhlh mutant livers, whereas inactivation of Hif-2α resulted in increased Pgk transcript levels, suggesting that Hif-1α is the predominant regulator of Hif-induced Pgk expression in vivo (Figure 3B). In contrast, the induction of Bnip3 in albumin-Vhlh mutant mice was not significantly decreased by inactivation of either Hif-1α or Hif-2α; however, inactivation of Arnt resulted in complete suppression, suggesting that both Hif-1α and Hif-2α have the capacity to regulate Bnip3 (Figure 3C). Based on these data, we conclude that Hif-1 and Hif-2 regulate the expression of distinct target genes in the liver. Importantly, our data demonstrate that hepatic Epo and Trf expression can be induced in the liver through the stabilization of Hif-2.
Hif-2 is required for the induction of hepatic Epo expression in anemic mice.
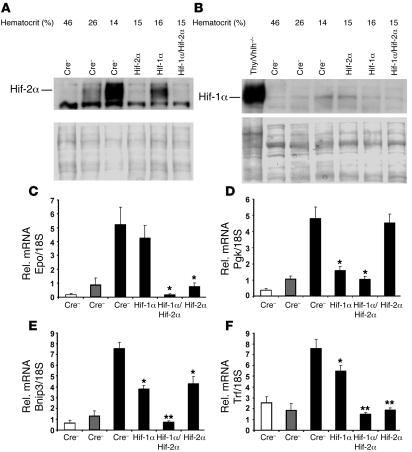
We next sought to determine whether Hif-2 is the dominant Hif in the regulation of hepatic Epo in a setting in which hepatocytes are WT for pVHL. We determined the relative contributions of Hif-1α and Hif-2α to hepatic Epo production mice with severe anemia induced by PHZ treatment and phlebotomy as an in vivo model of hepatic Hif stabilization and Epo activation. PHZ is a potent oxidant that causes severe hemolytic anemia resulting in increased Epo in the liver and kidney (36, 37). After induction of anemia in albumin-Cre mutant mice, blood was collected to monitor hematocrit values, and hepatic Epo transcript levels were assessed by real-time PCR. PHZ injection decreased hematocrit values in control mice from 45% ± 3% to 27% ± 3% within 40 hours after the initial injection (Figure 4A, first 2 lanes). Hematocrit values were reduced to 15% ± 2% following phlebotomy, which was sufficient to stabilize both Hif-1α and Hif-2α in the liver after 4 hours (Figure 4, A and B). Accordingly, we observed the greatest increase in hepatic Epo mRNA levels in mice with the lowest hematocrit values (Figure 4C). Inactivation of Hif-1α did not affect the induction in hepatic Epo despite a significant reduction in Pgk and Bnip3 mRNA levels (Figure 4, D and E). In contrast, inactivation of Hif-2α significantly reduced Epo, Bnip3, and Trf mRNA levels, whereas Pgk expression levels were unaffected. The combined deletion of Hif-1α and Hif-2α completely suppressed the induction of Epo, Pgk, Trf, and Bnip3 in acutely anemic mice (Figure 4, C–F). Thus, our data provide evidence that the hypoxic induction of hepatic Epo and Trf expression is preferentially mediated by Hif-2 and that the Hif-2 dominance in regulating Epo expression is not restricted to Vhlh-deficient hepatocytes.
Figure 4. Hepatic Epo expression in anemic mice is Hif-2α dependent.
(A and B) Hif-1α and Hif-2α proteins are stabilized in the livers of anemic mice as determined by Western blot analysis. Nuclear protein extracts were isolated from livers of albumin-Cre mutant mice of the indicated genotypes. Nuclear protein extracts isolated from the thymus (Thy) of Vhlh-deficient mice was used as a positive Hif-1α protein control. Ponceau S staining is shown to demonstrate equal protein loading. (C–F) Real-time PCR analysis of hepatic Epo, Pgk, Bnip3, and Trf expression in anemic albumin-Cre mutant mice. White bars represent mice with hematocrit values of approximately 45%; gray bars, approximately 27%; and black bars, approximately 15%. Bars represent average mRNA transcript levels of 3 mice for each group; error bars indicate SEM. *P < 0.05, **P < 0.001 compared with control mice (Cre–) with the same range of hematocrit as determined by Student’s t test.
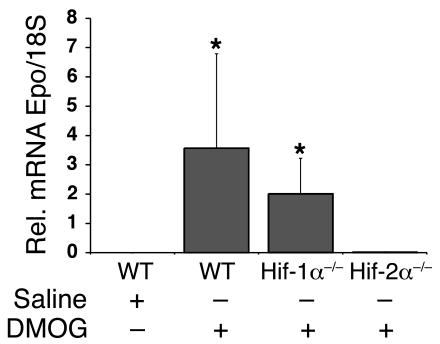
We next investigated whether hepatic Epo expression induced through nonhypoxic, pharmacological stabilization of Hif-α is Hif-2 dependent. Dimethyloxalylglycine (DMOG) has been previously shown to stabilize Hif-α through inhibition of HIF prolyl hydroxylation in vitro and in vivo (38). Eight-week-old albumin-Cre mutants or control littermates were injected i.p. with either isotonic saline or 10 mg of DMOG. Hepatic Epo mRNA levels were determined by real-time PCR 6 hours after injection. DMOG-treated Cre-negative control littermates and albumin–Hif-1α mutants showed a statistically significant increase in hepatic Epo mRNA levels compared with DMOG-treated albumin–Hif-2α mutants (both P = 0.0357 [Wilcoxon rank sum test]; n = 3 each). Epo levels in albumin–Hif-2α mutants were comparable to those in saline-injected animals (Figure 5; P = 0.1429). These results suggest that Hif-2 also regulates Epo under nonhypoxic conditions in Vhlh-competent hepatocytes.
Figure 5. Inhibition of Hif prolyl hydroxylation results in an Hif-2–dependent induction of hepatic Epo.
Relative Epo mRNA levels in the liver of albumin-Cre mutant mice treated with either with normal saline or 10 mg of DMOG as determined by real-time PCR. Bars represent the mean Epo mRNA transcript level of 3 mice for the WT and albumin–Hif-1α mutants (Hif-1α–/–) and 5 mice for the albumin-Hif-2α (Hif-2α–/–) mutants. Error bars represent SEM. *P < 0.05 compared with albumin–Hif-2α mutant mice as determined by the Wilcoxon rank sum test.
Hepatic Hif-2 regulates Epo during early postnatal development.
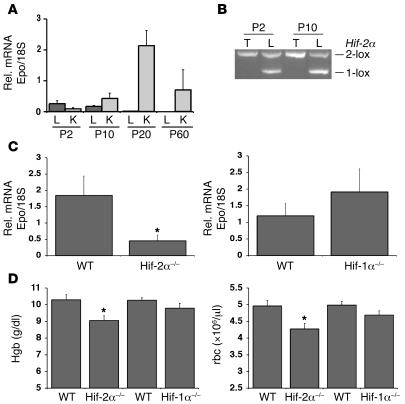
After we established that Hif-2α is required for the induction of hepatic Epo under pathological conditions, we next sought to define a physiological role for Hif-2–mediated Epo expression in the liver. Since hepatocytes are the major source of Epo during embryonic and early postnatal development (4–7), we investigated whether absence of Hif-2α would affect erythropoiesis in infant mice using albumin-Cre mutants. First, we examined hepatic Epo levels in WT postnatal mice by real-time PCR analysis and found that hepatic Epo was expressed in infant mice at P2 and P10 but not at P20 (Figure 6A). While hepatic Epo expression declined during early postnatal development, renal Epo levels increased, which is consistent with the switch in the primary source of systemic Epo from the liver to the kidney during this developmental period (Figure 6A; refs. 4–7). We next examined hepatic Hif-α protein levels in mice at P2, P4, and P10 by Western blot analysis. Both Hif-2α and Hif-1α were stable in nuclear extracts isolated from livers at P2, P4, and P8, whereas Hif-2α and Hif-1α were undetectable in P20 livers (data not shown). Since we observed that albumin-Cre–mediated recombination of Hif conditional alleles increased from P2 to P10 (Figure 6B), as previously observed by Postic et al. using a Cre-recombinase reporter allele (39), we determined liver Epo mRNA levels by real-time PCR in mice at P10. We found a 4-fold reduction in hepatic Epo levels in albumin–Hif-2α mutants compared with control, whereas liver Epo levels in albumin–Hif-1α mutants were not statistically significantly different from those in littermate controls (Figure 6C). The reduction in liver Epo levels in infant albumin–Hif-2α mutants was associated with a statistically significant decrease in hemoglobin concentrations (9.06 ± 0.29 g/dl versus 10.3 g/dl ± 0.3; P = 0.007) and red blood cell numbers (4.23 × 106 ± 0.16 × 106/μl versus 4.97 × 106 ± 0.16 × 106/μl; P = 0.006). In contrast, hemoglobin concentrations and red blood cell numbers did not statistically differ in adult albumin–Hif-2α mutants compared with control littermates (data not shown). Together, our data suggest that hepatic Hif-2α has a physiological role in the regulation of hepatic Epo expression and is important for the maintenance of erythropoiesis during infancy before the switch in EPO production from the liver to the kidney is completed.
Figure 6. Hif-2 regulates hepatic Epo expression in infant mice.
(A) Relative Epo mRNA levels in the livers and kidneys (K) of P2, P10, P20, and P60 mice. Bars represent the average Epo mRNA transcript level of 3 mice for each condition. (B) Genomic PCR analysis of DNA isolated from the tail and liver of albumin–Hif-2α mutant mice at P2 and P10. Note that the ratio of the recombined (1-lox) allele to the unrecombined (2-lox) Hif-2α allele increased between P2 and P10. (C) Relative Epo mRNA expression levels in P10 albumin–Hif-2α (Hif-2α–/–) and albumin–Hif-1α (Hif-1α–/–) and Cre-recombinase–negative control littermates (WT). Bars represent the mean Epo mRNA transcript level for WT albumin–Hif-2α littermates (n = 12) and albumin–Hif-2α mutants (n = 13) (left) and albumin–Hif-1α mutants and littermate controls (n = 10 each) (left). (D) Hemoglobin concentrations and red blood cell numbers in blood collected from 10-day-old albumin-Cre mutant and control mice. Bars represent the mean values for albumin–Hif-2α controls and mutants (n = 12 each), albumin–Hif-1α controls (n = 18), and albumin–Hif-1α mutants (n = 11). Error bars represent SEM. *P < 0.05 compared with littermate controls as determined by Student’s t test.
HIF-2 preferentially binds to the EPO 3' enhancer HRE in hepatocytes.
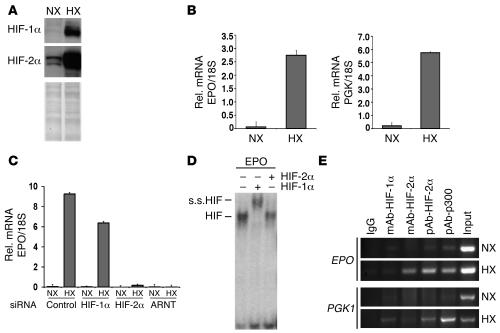
We next utilized an in vitro model to investigate the molecular mechanisms of HIF-2–mediated EPO expression in hepatocytes. Hypoxia treatment in Hep3B cells resulted in the stabilization of HIF-1α and HIF-2α and the transcriptional activation of EPO and PGK1 (Figure 7, A and B). Consistent with our results in vivo, the hypoxic induction of EPO in Hep3B cells was HIF-2α dependent and HIF-1α independent as determined by siRNA-mediated knockdown of HIF-1α, HIF-2α, and ARNT (Figure 7C). To investigate the molecular mechanism by which HIF-2 preferentially mediates the hypoxic induction of hepatic EPO, we initially examined the ability of HIF-1 and HIF-2 to bind to the EPO 3' enhancer HRE by gel shift analysis. The EPO 3' HRE is located within the liver inducibility element, which functions as a hypoxia-inducible enhancer in Hep3B cells (18, 40). Antibodies against HIF-1α, but not HIF-2α, supershifted the majority of the HIF/HRE complex, suggesting that HIF-1 preferentially binds to the unmodified EPO HRE (Figure 7D). Protein extracts from 786-0 cells were used as a positive control for the HIF-2α supershift (data not shown). We next examined the association of HIF-1α and HIF-2α with the endogenous EPO HRE in hepatocytes by chromatin immunoprecipitation (ChIP) analysis. Surprisingly, we observed that HIF-2α, but not HIF-1α, associated with the EPO HRE under hypoxia (Figure 7E). In contrast, the HRE of HIF-1 regulated PGK1 was bound by both HIF-1α and HIF-2α under hypoxic conditions, suggesting that HIF/HRE binding by itself is not sufficient to induce gene expression and that additional cofactors may be required for the induction of HIF target gene expression (Figure 7E). We conclude that HIF-2 mediates EPO expression in Hep3B cells as a result of a preferential HIF-2 interaction with the endogenous EPO 3′ HRE.
Figure 7. HIF-2α preferentially binds to the endogenous EPO 3′ HRE in hepatocytes.
(A) Western blot analysis for HIF-1 and HIF-2 in normoxic (21% O2; NX) and hypoxic (1% O2; HX) Hep3B cells. (B) EPO and PGK1 mRNA levels in normoxic and hypoxic Hep3B cells as determined by real-time PCR. (C) The hypoxic induction of EPO expression in Hep3B cells is HIF-2 dependent. Real-time PCR analysis of EPO expression in normoxic and hypoxic Hep3B cells treated with control, HIF-1α, HIF-2α, or control siRNA oligonucleotides. Error bars represent SD. (D) HIF-1α preferentially binds to the EPO HRE in vitro as determined by EMSA. s.s.HIF, HIF supershift. + and – indicate the presence and absence, respectively, of the antibody used in a supershift reaction. (E) ChIP analysis of the EPO and PGK1 HREs in normoxic and hypoxic Hep3B cells using antibodies directed against HIF-1α, HIF-2α, and CBP/p300. Coprecipitated DNA fragments were detected by PCR using primers spanning the EPO and PGK1 HREs. mAb-HIF-1α, mAb against HIF-1α; pAb-HIF-2α, polyclonal antibody against human HIF-2α; pAb-p300, polyclonal antibody against human p300.
Discussion
In the present study, we used a genetic approach to investigate the role of HIF-1 and HIF-2 in hepatic EPO production in adult and infant mice. We have shown that Hif-2 is the dominant Hif for the regulation of hepatic Epo expression in both Vhlh-deficient and anemic mice, whereas Hif-1 is required for the hypoxic regulation of other genes, such as Pgk. Furthermore, we have shown that Hif-2 during infancy is important for physiological liver Epo production and erythropoiesis.
The role of HIF-2 in the regulation of EPO expression does not appear to be restricted to hepatocytes. Studies in mice with germline-transmitted Hif-2α knockout or hypomorph alleles have suggested that renal Epo as well as retinal Epo production are dependent on Hif-2; however, the relative contributions of Hif-1 and Hif-2 could not be examined (41, 42). Germline inactivation of Hif-2α resulted in the development of anemia in adult mice that was found to be Epo dependent and associated with decreased renal Epo levels (41). In contrast, global inactivation of Hif-1α in the adult using a ubiquitously expressed inducible Cre-recombinase did not result in anemia (29). The kidney serves as the main physiologic oxygen sensor in the adult and responds to systemic hypoxia with a rapid increase in Epo production by cortical and outer medullary fibroblast-like renal interstitial cells (43). In the adult rodent kidney, Hif-2α, but not Hif-1α, was found to be strongly expressed in endothelial cells and in cortical renal interstitial fibroblasts under conditions of carbon monoxide treatment or renal ischemia, indirectly supporting the notion that Hif-2α may regulate Epo in renal interstitial cells (27, 44). However, a direct comparison of Hif-1α– and Hif-2α–deficient renal interstitial cells would be necessary to provide a definitive answer regarding the role of Hif-1 and Hif-2 in the regulation of renal Epo production. In this study, we have generated a mouse model that allows a direct comparison of Hif-1 to Hif-2 with regard to their ability to regulate hepatic Epo under physiologic conditions. Despite the technical limitations of albumin-Cre–mediated recombination in neonatal mice, we were able to show that hepatic Epo expression is Hif-2 dependent during infancy before the kidney becomes the main site of Epo production. The decrease in liver Epo levels in infant mice was associated with lower hematocrit values. In contrast, adult albumin–Hif-2α mutant mice did not develop anemia, supporting the notion that hepatocytes are the major source of systemic EPO during embryonic and early postnatal development but not during adulthood. Consistent with our observations, mutations in the Epo 3' HRE resulted in the development of neonatal anemia that resolves 2 weeks after birth (45). This finding also illustrates the importance of the EPO 3' HRE for hepatic EPO expression, as has been suggested by Semenza et al. (18). Whether additional Hif-2α–expressing cell types, such as cardiomyocytes, glial cells, and type II pneumocytes (27), are capable of producing Epo in an Hif-2–dependent manner remains to be investigated, as low levels of Epo mRNA have been detected in many different rat tissues including the lung, spleen, brain, and testis (46–48).
Our data demonstrate that HIF-1 and HIF-2 activate distinct target genes in vivo. Evidence that HIF-1 and HIF-2 have different functional roles was first observed when the phenotypes of Hif-1α– and Hif-2α–knockout mice were compared. Germline inactivation of Hif-1α results in embryonic lethality at E10 due to severe vascular and CNS defects (28, 49). In contrast, inactivation of Hif-2α by 3 independent groups resulted in embryonic lethality or death shortly after birth due to insufficient catecholamine synthesis, defects in vascular remodeling, or insufficient surfactant production from type II alveolar cells, depending on the genetic background (50–52). Inactivation of Hif-2α in a mixed 129/C57BL/6 genetic background resulted in viable mice that developed multiple-organ pathology associated with mitochondrial dysfunction (53). While some of the phenotypic differences observed between Hif-1α– and Hif-2α–knockout mice may be attributed to distinct Hif-1 and Hif-2 expression patterns, it seems likely that functional differences observed in cells expressing both HIF-1 and HIF-2 are a result of differential target gene activation (35). Although HIF-1α and HIF-2α are highly homologous in their DNA binding (83.9%) and dimerization (66.5%) domains, the remaining portion of the protein containing the N-terminal and C-terminal transactivation domains differ, with only 36.4% amino acid sequence homology, and may contribute to their unique functions (54).
Using a genetic approach, we were able to demonstrate that Hif-2, and not Hif-1, preferentially regulates hepatic Epo expression in vivo and propose that HIF-2 dominance in the regulation of hepatic EPO is mediated by the preferential association of HIF-2 with the endogenous EPO 3′ HRE. We observed by ChIP analysis that HIF-2, but not HIF-1, associates with the endogenous EPO 3′ HRE in hypoxic Hep3B cells. In contrast, we found that HIF-1 preferentially binds to the unmodified EPO HRE fragment in vitro, consistent with the purification of HIF-1 from hypoxic Hep3B extracts using an 18-nucleotide fragment containing the EPO HRE (11). Collectively, these data suggest that HIF-2 binding to the EPO 3' enhancer HRE requires additional nuclear factors associated with the EPO gene in vivo. This notion is supported by the recent finding that HIF-2–mediated expression of an EPO HRE–luciferase construct requires the minimal 223-bp enhancer fragment, which contains additional transcription factor binding sites (11, 19). Examination of the EPO 3' enhancer element has revealed 2 sites in addition to the HRE that are required for the hypoxic induction of EPO expression. One of these sites includes a DR-2 element that is a binding site for members of the nuclear hormone receptor family. It has been shown that hepatocyte nuclear factor–4 (HNF-4) binds to the EPO 3' enhancer DR-2 element, and it has been proposed as a candidate factor that may cooperate with HIF-2 (19). Similar to HIF-2, HNF-4 is expressed in a pattern that coincides with sites of Epo production in the liver and renal cortex and is required for the hypoxic induction of EPO expression in Hep3B cells (19, 55, 56). The notion that HIF-2 target gene selectivity depends on the availability and cooperation of HIF with transcription factors bound to cognate elements in target gene regulatory sequences has been previously suggested; however, further studies will need to be performed to isolate factors that interact with HIF-2 and mediate the hypoxic induction of EPO (57).
The ability of EPO-producing cells to efficiently target HIF-α subunits for proteasomal degradation under normoxia is essential for normal erythropoiesis and has clinical importance. Patients with congenital Chuvash polycythemia are homozygous for the Arg200Trp mutation in the VHL tumor suppressor and have elevated red blood cell counts due to increased EPO production as a result of elevated HIF activity (58, 59). Furthermore, mutations in PHD-2, the most abundant PHD protein, result in a rare form of familial erythrocytosis (with hematocrit values up to 54%) from an inability to properly interact with HIF-α under normoxia (60). Our data suggest that inhibition of HIF-α degradation in the liver is sufficient to substantially raise systemic Epo levels (up to 40-fold in Vhlh mutants; ref. 32) and thus may be useful for the treatment of anemia. The pharmacological stabilization of HIF-2 in the liver could potentially be achieved through the oral administration of specific HIF prolyl hydroxylase inhibitors. Individual PHD isoforms (PHD1, PHD2, and PHD3) have the ability to differentially regulate HIF-1α and HIF-2α stability (61). Thus, one could imagine that compounds targeting individual PHD isoforms with different efficiencies could shift the HIF-1α/HIF-2α ratio toward HIF-2α. Furthermore, pharmacological targeting of HIF-α degradation by prolyl hydroxylase inhibition appears to preferentially induce plasma Epo levels in bilateral nephrectomized mice (62), as well as increase serum EPO levels in humans (63), and improve anemia of chronic disease and inflammation in animal models (64). In the latter studies, inhibition of HIF-α hydroxylation not only increased serum EPO levels but also improved iron uptake and metabolism. Thus, it is not surprising that Trf and its receptor have previously been found to be regulated by HIF (22, 33, 65, 66). Our studies indicate that in addition to that of Epo, expression of Trf is preferentially regulated by Hif-2 in the liver, suggesting that HIF-2 through its potential effects on iron homeostasis may play a broader role in the regulation of erythropoiesis. The role of HIF-2 in the hypoxic regulation of additional genes involved in iron metabolism awaits future investigation.
In summary, we have shown by direct genetic comparison that HIF-1 and HIF-2 have distinct functions with regard to hypoxic gene regulation in the liver. We demonstrate that hepatic Epo expression is preferentially regulated by Hif-2 and not Hif-1 in the early postnatal, Vhlh-deficient, and anemic adult liver. The finding that HIF-1 and HIF-2 are not interchangeable with regard to the regulation of EPO has implications for pharmacological strategies that aim at targeting the HIF oxygen-sensing pathway for the treatment of anemia.
Methods
Generation and analysis of mice.
The generation of Vhlh, Hif-1α, Hif-2α, and Arnt conditional alleles and the albumin-Cre and PEPCK-Cre transgenes has been previously described (29, 30, 32, 67–69). Cre-mediated inactivation of pVHL, HIF-1α, HIF-2α, and ARNT in hepatocytes was accomplished by generating mice that were homozygous for the respective 2-lox alleles and expressed either the PEPCK-Cre or albumin-Cre transgene. The genotypes were as follows: (a) [Vhlh2lox/2lox;PEPCK-Cre] and [Vhlh2lox/2lox;albumin-Cre], referred to as PEPCK-Vhlh or albumin-Vhlh mutant; (b) [Vhlh2lox/2loxHif1a2lox/2lox;albumin-Cre], referred to as albumin-Vhlh/Hif-1α mutant; (c) [Vhlh2lox/2loxEpas12lox/2lox;PEPCK-Cre] and [Vhlh2lox/2loxEpas12lox/2lox; albumin-Cre], referred to as PEPCK-Vhlh/Hif-2α or albumin-Vhlh/Hif-2α mutant; (d) [Vhlh2lox/2loxArnt3lox/3lox;albumin-Cre], referred to as albumin-Vhlh/Arnt mutant; (e) [Vhlh2lox/2loxHif1a2lox/2loxEpas12lox/2lox;albumin-Cre], referred to albumin-Vhlh/Hif-1α/Hif-2α mutant; (f) [Hif1a2lox/2lox;albumin-Cre], referred to albumin–Hif-1α mutant; and (g) [Epas12lox/2lox;albumin-Cre], referred to albumin–Hif-2α mutant.
Littermates not carrying the Cre transgene were used as control animals. Mutant mice were generated in a mixed genetic background (BALB/c, 129Sv/J, C57BL/6). The primer sequences used to detect 2-lox, 1-lox, and the WT alleles for Vhlh, Hif-1α, and Hif-2α have been previously described (29, 68).
To induce anemia, 2-month-old mice were subcutaneously injected with 40 mg/kg of PHZ hydrochloride (0.4% in normal saline, pH 7.2) 3 times at 12-hour intervals. Forty hours after the initial injection, mice were anesthetized, and approximately 30%–40% of total blood volume was removed by retroorbital bleeding and was replaced by twice the amount of sterile normal saline by i.p. injection. Mice were kept under complete anesthesia on a heating pad for 4 hours, and the livers were harvested after euthanization. For DMOG treatment, 10 mg DMOG was dissolved in 0.5 ml of isotonic saline and injected i.p. into 8-week-old female mice.
All procedures involving mice were performed in accordance with the NIH guidelines for use and care of live animals and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC).
For blood analysis, hematocrit values, red blood cell numbers, and hemoglobin concentrations were determined in blood using a Hemavet 1500 CBC analyzer (CDC Technologies). Hematocrit values were also determined by centrifuging blood in microcapillary tubes.
DNA, RNA, and protein isolation.
Mouse tail DNA was isolated according to Laird et al. and used for genomic PCR (70). DNA from mouse tissues was isolated using TRI zol reagent according to the manufacturer’s instructions (Invitrogen). RNA was isolated using RNeasy Maxi Kit according to the manufacturer’s protocols (QIAGEN).
Nuclear protein extracts were prepared and analyzed by Western blot as previously described (24). Primary antibodies for Western blot analysis include a polyclonal Hif-2α antibody (raised against amino acids 580–693) and a monoclonal Hif-1α antibody (Novus Biologicals).
Gel shift.
The EPO sense (5'-GCCCTACGTGCTGTCTCACACAGC-3') and antisense oligonucleotides (5'-GCTGTGTGAGACAGCACGTA-3') were annealed and subject to EMSA as previously described (24). Antibodies used for supershift reactions were the monoclonal HIF-1α (NB100-105; Novus Biologicals) and HIF-2α (NB100-132; Novus Biologicals) antibodies. As a positive control for the HIF-2 supershift, nuclear extracts from the HIF-2–expressing cell line 786-0 were included (data not shown).
RT-PCR.
cDNA was synthesized from 4 μg of DNase (Invitrogen) treated RNA using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). One microliter of cDNA was subjected to PCR amplification using either SYBR Green PCR Master Mix or Taqman Universal PCR Master Mix (both from Applied Biosystems). The following primer and probe sets were used to amplify specific target genes: SYBR Green primers: Trf (forward, 5'-TGGAGACAGATGCTCCCTCC-3'; reverse, 5'-TTTGTGCTCTGTGTATGTGGTAAGG-3'); primers for Epo, Pgk, Bnip3, and 18S were previously published (24).
PCR amplification was performed on the ABI Prism 7000 Sequence Detection System (Applied Biosystems) as previously described (24). 18S was used to normalize mRNA. Relative quantitation of mRNA expression levels was determined using the relative standard curve method according to the manufacturer’s instructions (Applied Biosystems).
RNA interference studies.
Hep3B cells were transfected with 5 nM of control, HIF-1α, or HIF-2α siRNA oligonucleotides using the HiPerFect Transfection Reagent according to the manufacturer’s protocols (QIAGEN). Forty-eight hours after transfection, hypoxic samples were exposed to 1% oxygen for 16 hours. siRNA oligonucleotides were purchased through QIAGEN’s pre-validated siRNA database: ARNT siRNA target sequence, GAAGUCAGAUGGUUUAUUU; HIF-1 HP validated siRNA, catalog SI02664053; HIF-2 HP validated siRNA, catalog SI02663038. Primers used for real-time PCR analysis were: EPO (forward, 5'-ACCAACATTGCTTGTGCCAC-3'; reverse, 5'-TCTGAATGCTTCCTGCTCTGG-3').
ChIP assay.
The ChIP assay was performed according to the protocol provided with the ChIP assay kit (Upstate). Briefly, Hep3B cells were exposed to hypoxia (1.0% O2) overnight and incubated with 1 μM of DMOG prior to harvest. The primary antibodies used for immunoprecipitation included: anti–HIF-1α (NB100-105; Novus Biologicals Inc.), anti–HIF-2α (NB100-132; Novus Biologicals Inc.), anti–HIF-2α (NB100-122; Novus Biologicals Inc.), and anti-p300 (N-15 and C-20; Santa Cruz Biotechnology Inc.). The primer sequences, designed to amplify the HRE-containing region of the EPO and PGK promoters, used were: EPO forward primer (5'-TCGTTTTCTGGGAACCTCCA-3') and EPO reverse primer (5'-GGAGCCACCTTATTGACCAG-3'); PGK forward primer (5'-GTTCGCAGC GTCACCCGGATCTTCG-3') and PGK reverse primer (5'-AGGCTTGCAGAATGCGGAACACC-3').
Statistics.
Statistical analysis was performed using the unpaired 2-tailed Student’s t test and the Wilcoxon rank sum test. P values of less than 0.05 were considered significant.
Acknowledgments
This work was supported by NIH grant DK073467 and in part by NIH grant CA100787 (both to V.H. Haase) and by the Center for Molecular Studies in Digestive and Liver Disease (P30-DK50306). E.B. Rankin is supported by a fellowship grant awarded by the American Heart Association. The authors would like to thank Debra Higgins for critical review of the manuscript.
Footnotes
Nonstandard abbreviations used: ARNT, aryl-hydrocarbon-receptor nuclear translocator; bHLH, basic helix-loop-helix; Bnip3, BCL2/adenovirus E1B–interacting protein 1, NIP3; ChIP, chromatin immunoprecipitation; DMOG, dimethyloxalylglycine; EPO, erythropoietin; HIF-1, hypoxia-inducible factor–1; HRE, hypoxia response element; PGK, phosphoglycerate kinase 1; PHD, prolyl-4-hydroxylase domain; PHZ, phenylhydrazine; Trf, transferrin; VHL, von Hippel–Lindau; Vhlh, VHL syndrome homolog.
Conflict of interest: V.H. Haase declares that he receives research grant support from GlaxoSmithKline. M.P. Biju is now an employee of GlaxoSmithKline.
Citation for this article: J. Clin. Invest. 117:1068–1077 (2007). doi:10.1172/JCI30117
Erinn B. Rankin and Mangatt P. Biju contributed equally to this work.
See the related Commentary beginning on page 862.
References
- 1.Ebert B.L., Bunn H.F. Regulation of the erythropoietin gene. Blood. 1999;94:1864–1877. [PubMed] [Google Scholar]
- 2.Wu H., Lee S.H., Gao J., Liu X., Iruela-Arispe M.L. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development. 1999;126:3597–3605. doi: 10.1242/dev.126.16.3597. [DOI] [PubMed] [Google Scholar]
- 3.Wu H., Liu X., Jaenisch R., Lodish H.F. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- 4.Zanjani E.D., Ascensao J.L., McGlave P.B., Banisadre M., Ash R.C. Studies on the liver to kidney switch of erythropoietin production. J. Clin. Invest. 1981;67:1183–1188. doi: 10.1172/JCI110133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dame C., et al. Erythropoietin mRNA expression in human fetal and neonatal tissue. Blood. 1998;92:3218–3225. [PubMed] [Google Scholar]
- 6.Moritz K.M., Lim G.B., Wintour E.M. Developmental regulation of erythropoietin and erythropoiesis. Am. J. Physiol. 1997;273:R1829–R1844. doi: 10.1152/ajpregu.1997.273.6.R1829. [DOI] [PubMed] [Google Scholar]
- 7.Eckardt K.U., Ratcliffe P.J., Tan C.C., Bauer C., Kurtz A. Age-dependent expression of the erythropoietin gene in rat liver and kidneys. J. Clin. Invest. 1992;89:753–760. doi: 10.1172/JCI115652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koury S.T., Bondurant M.C., Koury M.J., Semenza G.L. Localization of cells producing erythropoietin in murine liver by in situ hybridization. Blood. 1991;77:2497–2503. [PubMed] [Google Scholar]
- 9.Fandrey J. Oxygen-dependent and tissue-specific regulation of erythropoietin gene expression. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;286:R977–R988. doi: 10.1152/ajpregu.00577.2003. [DOI] [PubMed] [Google Scholar]
- 10.Eckardt K.U., Kurtz A. Regulation of erythropoietin production. Eur. J. Clin. Invest. 2005;35(Suppl. 3):13–19. doi: 10.1111/j.1365-2362.2005.01525.x. [DOI] [PubMed] [Google Scholar]
- 11.Semenza G.L., Wang G.L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G.L., Semenza G.L. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 14.Kewley R.J., Whitelaw M.L., Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 15.Wenger R.H. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- 16.Semenza G.L. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 17.Schofield C.J., Ratcliffe P.J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 18.Semenza G.L., Koury S.T., Nejfelt M.K., Gearhart J.D., Antonarakis S.E. Cell-type-specific and hypoxia-inducible expression of the human erythropoietin gene in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8725–8729. doi: 10.1073/pnas.88.19.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warnecke C., et al. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J. 2004;18:1462–1464. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 20.Stolze I., et al. Hypoxia-inducible erythropoietin gene expression in human neuroblastoma cells. Blood. 2002;100:2623–2628. doi: 10.1182/blood-2001-12-0169. [DOI] [PubMed] [Google Scholar]
- 21.Tam B.Y., et al. VEGF modulates erythropoiesis through regulation of adult hepatic erythropoietin synthesis. Nat. Med. 2006;12:793–800. doi: 10.1038/nm1428. [DOI] [PubMed] [Google Scholar]
- 22.Yoon D., et al. HIF-1alpha-deficiency results in dysregulated EPO signaling and iron homeostasis in mouse development. J. Biol. Chem. 2006;281:25703–25711. doi: 10.1074/jbc.M602329200. [DOI] [PubMed] [Google Scholar]
- 23.Grimm C., et al. HIF-1-induced erythropoietin in the hypoxic retina protects against light-induced retinal degeneration. Nat. Med. 2002;8:718–724. doi: 10.1038/nm723. [DOI] [PubMed] [Google Scholar]
- 24.Rankin E.B., et al. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol. Cell. Biol. 2005;25:3163–3172. doi: 10.1128/MCB.25.8.3163-3172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kietzmann T., Cornesse Y., Brechtel K., Modaressi S., Jungermann K. Perivenous expression of the mRNA of the three hypoxia-inducible factor alpha-subunits, HIF1alpha, HIF2alpha and HIF3alpha, in rat liver. Biochem. J. 2001;354:531–537. doi: 10.1042/0264-6021:3540531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroka D.M., et al. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 2001;15:2445–2453. doi: 10.1096/fj.01-0125com. [DOI] [PubMed] [Google Scholar]
- 27.Wiesener M.S., et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 28.Ryan H.E., Lo J., Johnson R.S. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber M., et al. Acute postnatal ablation of Hif-2alpha results in anemia. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postic C., et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 31.Kaelin W.G., Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 32.Haase V.H., Glickman J.N., Socolovsky M., Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolfs A., Kvietikova I., Gassmann M., Wenger R.H. Oxygen-regulated transferrin expression is mediated by hypoxia-inducible factor-1. J. Biol. Chem. 1997;272:20055–20062. doi: 10.1074/jbc.272.32.20055. [DOI] [PubMed] [Google Scholar]
- 34.Hu C.J., Wang L.Y., Chodosh L.A., Keith B., Simon M.C. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol. Cell. Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raval R.R., et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Socolovsky M., et al. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 37.Criswell K.A., Sulkanen A.P., Hochbaum A.F., Bleavins M.R. Effects of phenylhydrazine or phlebotomy on peripheral blood, bone marrow and erythropoietin in Wistar rats. J. Appl. Toxicol. 2000;20:25–34. doi: 10.1002/(sici)1099-1263(200001/02)20:1<25::aid-jat624>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 38.Milkiewicz M., Pugh C.W., Egginton S. Inhibition of endogenous HIF inactivation induces angiogenesis in ischaemic skeletal muscles of mice. J. Physiol. 2004;560:21–26. doi: 10.1113/jphysiol.2004.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postic C., Magnuson M.A. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 40.Semenza G.L., Nejfelt M.K., Chi S.M., Antonarakis S.E. Hypoxia-inducible nuclear factors bind to an enhancer element located 3’ to the human erythropoietin gene. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scortegagna M., et al. HIF-2alpha regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood. 2005;105:3133–3140. doi: 10.1182/blood-2004-05-1695. [DOI] [PubMed] [Google Scholar]
- 42.Morita M., et al. HLF/HIF-2alpha is a key factor in retinopathy of prematurity in association with erythropoietin. EMBO J. 2003;22:1134–1146. doi: 10.1093/emboj/cdg117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maxwell P.H., et al. Identification of the renal erythropoietin-producing cells using transgenic mice. Kidney Int. 1993;44:1149–1162. doi: 10.1038/ki.1993.362. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberger C., et al. Expression of hypoxia-inducible factor-1alpha and -2alpha in hypoxic and ischemic rat kidneys. J. Am. Soc. Nephrol. 2002;13:1721–1732. doi: 10.1097/01.asn.0000017223.49823.2a. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki N., et al. Regulation and essential role of the erythropoietin gene in the liver [abstract]. Ann. Hematol. 2006;85:656. [Google Scholar]
- 46.Tan C.C., Eckardt K.U., Ratcliffe P.J. Organ distribution of erythropoietin messenger RNA in normal and uremic rats. Kidney Int. 1991;40:69–76. doi: 10.1038/ki.1991.181. [DOI] [PubMed] [Google Scholar]
- 47.Fandrey J., Bunn H.F. In vivo and in vitro regulation of erythropoietin mRNA: measurement by competitive polymerase chain reaction. Blood. 1993;81:617–623. [PubMed] [Google Scholar]
- 48.Marti H.H., et al. Erythropoietin gene expression in human, monkey and murine brain. Eur. J. Neurosci. 1996;8:666–676. doi: 10.1111/j.1460-9568.1996.tb01252.x. [DOI] [PubMed] [Google Scholar]
- 49.Iyer N.V., et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian H., Hammer R.E., Matsumoto A.M., Russell D.W., McKnight S.L. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng J., Zhang L., Drysdale L., Fong G.H. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Compernolle V., et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat. Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- 53.Scortegagna M., et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-/- mice. Nat. Genet. 2003;35:331–340. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- 54.Ema M., et al. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanchard K.L., Acquaviva A.M., Galson D.L., Bunn H.F. Hypoxic induction of the human erythropoietin gene: cooperation between the promoter and enhancer, each of which contains steroid receptor response elements. Mol. Cell. Biol. 1992;12:5373–5385. doi: 10.1128/mcb.12.12.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galson D.L., et al. The orphan receptor hepatic nuclear factor 4 functions as a transcriptional activator for tissue-specific and hypoxia-specific erythropoietin gene expression and is antagonized by EAR3/COUP-TF1. Mol. Cell. Biol. 1995;15:2135–2144. doi: 10.1128/mcb.15.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aprelikova O., Wood M., Tackett S., Chandramouli G.V., Barrett J.C. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 2006;66:5641–5647. doi: 10.1158/0008-5472.CAN-05-3345. [DOI] [PubMed] [Google Scholar]
- 58.Ang S.O., et al. Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat. Genet. 2002;32:614–621. doi: 10.1038/ng1019. [DOI] [PubMed] [Google Scholar]
- 59.Pastore Y., et al. Mutations of von Hippel-Lindau tumor-suppressor gene and congenital polycythemia. Am. J. Hum. Genet. 2003;73:412–419. doi: 10.1086/377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Percy M.J., et al. A family with erythrocytosis establishes a role for prolyl hydroxylase domain protein 2 in oxygen homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:654–659. doi: 10.1073/pnas.0508423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Appelhoff R.J., et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 62.Wang Q.J., et al. Induction of renal and extra-renal erythropoietin production by orally bioavailable HIF prolyl hydroxylase inhibitors [abstract]. J. Am. Soc. Nephrol. 2005;16:483A. [Google Scholar]
- 63.Liu D.Y., et al. Novel and beneficial pharmacodynamic properties of endogenous EPO and complete erythropoiesis induced by selective HIF prolyl hydroxylase inhibitors [abstract]. J. Am. Soc. Nephrol. 2005;16:761A. [Google Scholar]
- 64.Langsetmo I., et al. FG-2216 corrects anemia and improves iron utilization in a rat model of anemia of chronic disease: comparison to darbepoetin [abstract]. J. Am. Soc. Nephrol. 2005;16:481A. [Google Scholar]
- 65.Lok C.N., Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J. Biol. Chem. 1999;274:24147–24152. doi: 10.1074/jbc.274.34.24147. [DOI] [PubMed] [Google Scholar]
- 66.Tacchini L., Bianchi L., Bernelli-Zazzera A., Cairo G. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J. Biol. Chem. 1999;274:24142–24146. doi: 10.1074/jbc.274.34.24142. [DOI] [PubMed] [Google Scholar]
- 67.Ryan H.E., et al. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- 68.Rankin E.B., Tomaszewski J.E., Haase V.H. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walisser J.A., Bunger M.K., Glover E., Harstad E.B., Bradfield C.A. Patent ductus venosus and dioxin resistance in mice harboring a hypomorphic Arnt allele. J. Biol. Chem. 2004;279:16326–16331. doi: 10.1074/jbc.M400784200. [DOI] [PubMed] [Google Scholar]
- 70.Laird P.W., et al. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]