Abstract
The deregulation of homeobox (HOX) genes in acute myeloid leukemia (AML) and the potential for these master regulators to perturb normal hematopoiesis is well established. To date, overexpression of HOX genes in AML has been attributed to specific chromosomal aberrations and abnormalities involving mixed-lineage leukemia (MLL), an upstream regulator of HOX genes. The finding reported in this issue of the JCI by Scholl et al. that caudal-type homeobox transcription factor 2 (CDX2), which is capable of affecting HOX gene expression during embryogenesis, is overexpressed in 90% of patients with AML and induces a transplantable AML in murine models provides an alternative mechanism for HOX-induced leukemogenesis and yields important insights into the hierarchy of HOX gene regulation in AML (see the related article beginning on page 1037).
Acute myeloid leukemia (AML) is a heterogeneous disease in which hematopoietic progenitor cells acquire genetic lesions that lead to a block in differentiation, increased self-renewal, and unregulated proliferation. The emergence of leukemic blasts appears to require at least two major genetic “hits,” involving perturbations in growth factor signaling pathways and hematopoietic differentiation programs (1).
Among the receptor tyrosine kinases (RTKs), fms-like tyrosine kinase 3 (FLT3), which plays important roles in hematopoietic progenitor cell survival and proliferation, is overexpressed in a significant proportion of AMLs, and mutations resulting in the constitutive activation of FLT3 occur in approximately 33% of patients (2). Mutations leading to the constitutive activation of a related RTK, c-KIT, and of signaling intermediates such as RAS, are also frequently described in AML (3). The dysregulation of associated signaling pathways (e.g., Ras/MAPK, PI3K/AKT, and JAK/STAT) is thought to result in growth factor–independent proliferation and clonal expansion of hematopoietic progenitors.
The second hit targets transcription factors capable of disrupting hematopoietic cell differentiation. This may occur following the dysregulation of specific gene regulators as a result of gene amplification (e.g., v-myc myelocytomatosis viral oncogene homolog [MYC], mixed-lineage leukemia [MLL], genes at the chromosome 11q23 locus) (4); point mutations in transcriptional regulators (e.g., CCAAT/enhancer–binding protein [C/EBP], runt-related transcription factor 1 [RUNX1]); and chromosomal translocations resulting in the fusion of promyelocytic leukemia (PML) and the retinoic acid receptor α (RARa) to yield the PML-RARα chimeric protein or fusion of RUNX1 and runt-related transcription factor 1, translocated to 1 (RUNX1T1), which gives rise to the RUNX1-RUNX1T1 fusion protein (1). It is becoming increasingly clear that one set of genes commonly affected by these chimerical and mutated transcriptional regulators are the homeobox (HOX) genes. Accordingly, overexpression of homeobox master transcription factors, which fulfill critical roles in embryonic development, organogenesis, and normal hematopoietic differentiation, is a common feature of AML (5).
HOX genes: from hematopoiesis to leukemia
In mammals, HOX genes are located in two main clusters, the primordial cluster and the ParaHox cluster, which are thought to originate from the duplication of a hypothetical ProtoHox cluster of four genes early in evolution (6). The primordial HOX cluster consists of 13 paralogous groups of genes that exist as distinct, unlinked complexes on human chromosomes 7p15 (HOXA), 17q21 (HOXB), 12q13 (HOXC), and 2q31 (HOXD), and the cluster is organized such that during embryonic development, the order of expression along the anterior-posterior embryonic axis (3' to 5') is colinear with the alignment of genes on the chromosome (Figure 1). During hematopoiesis, HOX genes are expressed in lineage- and stage-specific combinations; however, cell commitment to myeloid or erythroid lineages is accompanied by global downregulation of HOX gene expression (7).
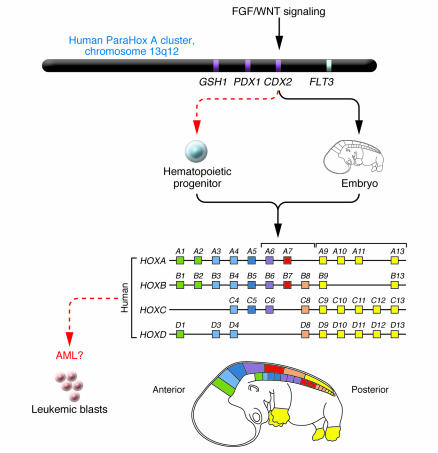
Figure 1. A model for CDX2-mediated leukemogenesis.
CDX2 is expressed in the posterior primitive streak during embryogenesis, where it directs anteroposterior axial development and elongation by regulating HOX gene expression. A study in this issue of the JCI (12) reports that monoallelic expression of CDX2 is observed in 90% of patients with AML and may perturb hematopoiesis by affecting HOX gene expression (red dashed arrows indicate ectopic expression; black arrows indicate normal expression in embryonic development). GSH1, GS homeobox 1; PDX1, pancreatic and duodenal homeobox 1. Figure modified with permission from Molecular Genetics and Metabolism (21).
HOX genes may be dysregulated in AML by several different mechanisms. First, specific HOX genes can be disrupted via chromosomal translocation. Specifically, HOXA9 and HOXD13 are dysregulated through the t(7;11) and t(2;11) translocations, respectively (8), in both cases creating fusion proteins between the HOX protein and the nucleoporin 98 kDa (NUP98) nuclear protein. Overexpression of HOXA6, HOXA7, HOXA9, and the HOX cofactor myeloid ecotropic viral integration site 1 (MEIS1) has also been correlated with chromosome 11q23 abnormalities involving the MLL protein, which regulates the expression of HOX genes (9). A less frequently observed translocation in AML, t(8;16)(p11;p13), which results in the overexpression of the MYST3-CREBBP fusion protein, is also associated with overexpression of HOXA9, HOXB9, HOXA10, and MEIS1 (10). In addition, the expression of HOXC4 was shown to be upregulated in the NB4 PML-RARα cell line following all-trans retinoic acid–induced (ATRA-induced) differentiation as well as in bone marrow from acute PML patients during ATRA treatment, strengthening the theory that dysregulated HOX expression is a signature feature of AML (11).
CDX2 overexpression: a unifying theme in AML?
In keeping with the theme of dysregulated HOX gene expression in AML, in this issue of the JCI Scholl et al. (12) demonstrate that caudal-type homeobox transcription factor 2 (CDX2), a homeobox gene involved in anteroposterior axis definition and intestinal epithelial cell differentiation (13), but strikingly absent in hematopoietic progenitors, is overexpressed in 90% of patients with AML. The leukemogenic potential of CDX2 overexpression was first described by Chase et al. (14) following the identification of a novel chromosomal rearrangement, t(12;13)(p13;q12), in a patient with AML, which yielded the ets variant gene 6–CDX2 (ETV6-CDX2) fusion protein in addition to full-length CDX2. Significantly, transduction of murine hematopoietic progenitors with CDX2, but not the ETV6-CDX2 fusion protein, resulted in a transplantable and fatal AML. Although ETV6-CDX2 induced myeloproliferation, it did not induce leukemia or accelerate CDX2-mediated leukemogenesis (15), suggesting that the primary oncogenic affect of this translocation is the ectopic expression of CDX2 in the hematopoietic compartment (Figure 1).
CDX2 belongs to the ParaHox A cluster, which consists of a three-gene complex including GS homeobox 1 (GSH1), pancreatic and duodenal homeobox 1 (PDX1), and CDX2 that exists on chromosome 13q12 (6) (Figure 1). On the basis of sequence similarity, CDX is more related to the posterior group of HOX genes, which includes paralogous groups 9–13, and during embryonic development is expressed in the posterior primitive streak to regulate HOX genes and in posterior signaling pathways to direct anteroposterior axis development and elongation. Given the role of CDX2 as a regulator of HOX genes in development, it is therefore not surprising that ectopic expression of CDX2 in hematopoietic progenitors perturbs blood cell differentiation programs strictly regulated by HOX genes. The finding by Scholl et al. (12) that overexpression of Cdx2 in primary murine hematopoietic progenitors resulted in transplantable AML in vivo, coinciding with a three-fold upregulation in Hoxb6 expression, is particularly intriguing given that overexpression of HOXB6 has been documented in approximately 40% of human AMLs that do not have chromosomal translocations (16). Identifying whether Cdx2 directly regulates Hoxb6 and providing evidence of the specific involvement of Hoxb6 in Cdx2-mediated leukemogenesis would strengthen the hypothesis that dysregulated HOX gene expression by CDX2 is a major pathway contributing to leukemogenesis.
The dysregulation of HOX genes by members of the caudal HOX family may prove to be a unifying theme in AML, particularly in light of a recent study by Bansal et al. (17) that revealed dysregulated CDX4 expression in 25% of AMLs. However, whereas CDX2 induced rapid AML in bone marrow transplant recipients, CDX4 induced a partially penetrant, long-latency AML, suggesting that these transcription factors regulate distinct sets of genes, including potentially different subsets of HOX genes. In addition to dysregulating HOX genes, CDX2 overexpression may also affect signaling pathways. Cdx1/2 mutant embryos demonstrate posterior body truncations that phenocopy loss-of-function Wnt3a and Fgfr1 mutants (18), suggesting an overlap among Cdx, Wnt, and Fgf pathways in axial patterning. This implies that some CDX2 target genes might include components of signaling pathways or that CDX2 itself may be a target of cell-signaling regulators, a point highlighted by Scholl et al. (12), given that related family members CDX1 and CDX4 are direct targets of the Wnt pathway commonly activated in AML. Indeed, Scholl et al. showed that knockdown of CDX2 slowed cellular proliferation specifically in CDX2-expressing AML cell lines. Taken together, these data suggest that gain-of-function mutations of CDX2 might augment cell signaling in addition to disrupting differentiation programs. This may explain the rapid onset of AML in mice transplanted with CDX2-expressing hematopoietic progenitors.
Unraveling the mechanism
The molecular basis for the overexpression of CDX2 remains elusive, and the absence of chromosomal rearrangements or mutations in the coding sequence or proximal promoter of CDX2 suggest that other undefined regulatory regions driving CDX2 expression may be a target for mutation. Indeed, aberrant expression of CDX2 in the absence of genetic abnormalities on chromosome 13 was previously documented in a case of chronic myeloid leukemia blast crisis (14). Intriguingly, microarray analysis of 50 AML patient specimens revealed that the highest levels of HOX expression are associated with a subset of patients with intermediate cytogenetics and elevated levels of FLT3 mRNA and FLT3 mutations. In the absence of mutations or amplifications, overexpression of FLT3 was correlated with aberrant expression of multiple HOX genes, suggesting that deregulation of HOX genes may be due to aberrant growth factor signaling (19). Given the close proximity of the FLT3 gene to the CDX2 gene and the fact that both proteins are overexpressed in a high percentage of AMLs, it is tempting to speculate that cis-acting activating mutations in promoter or enhancer elements in the vicinity of these genes may contribute to the leukemic initiation and progression and may account for the link among FLT3, CDX2, and HOX gene overexpression in the absence of detectable cytogenetics (20). Detailed analyses of the regulatory sequences flanking CDX2 and FLT3 could provide insights into the role of these oncogenes in AML. It would also be important to correlate the level of CDX2 overexpression in AML with HOX gene expression.
Identification of a HOX gene pathway disrupted in a substantial proportion of patients with AML (12) is a major clue to disease pathogenesis; however, more direct evidence of a link between CDX2 overexpression and the dysregulation of specific HOX genes is needed. For example, demonstration by chromatin precipitation that CDX2 occupies the promoters of genes such as HOXB8 and HOXB6 in CDX2-positive AML cell specimens would reinforce the importance of CDX2 as an aberrant regulator of HOX genes. In addition, identifying the mechanism by which CDX2 is overexpressed is crucial for unraveling the hierarchy of genetic pathways required for leukemia initiation and progression. While specific targeting of transcriptional regulators remains elusive, better understanding of the circuitry of the HOX genes in AML could present a new set of genetic targets, with potential therapeutic benefit for multiple AML subtypes.
Footnotes
Nonstandard abbreviations used: AML, acute myeloid leukemia; ATRA, all-trans retinoic acid; CDX2, caudal-type homeobox transcription factor 2; ETV6, ets variant gene 6; FLT3, fms-like tyrosine kinase 3; HOX, homeobox; MEIS1, myeloid ecotropic viral integration site 1; MLL, mixed-lineage leukemia; PML, promyelocytic leukemia; RARα, retinoic acid receptor α; RTK, receptor tyrosine kinase; RUNX1, runt-related transcription factor 1.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:865–868 (2007). doi:10.1172/JCI31861.
See the related article beginning on page 1037.
References
- 1.Licht J.D., Sternberg D.W. The molecular pathology of acute myeloid leukemia. Hematology Am. Soc. Hematol. Educ. Program. 2005;2005:137–142. doi: 10.1182/asheducation-2005.1.137. [DOI] [PubMed] [Google Scholar]
- 2.Small D. FLT3 mutations: biology and treatment. Hematology Am. Soc. Hematol. Educ. Program. 2006;2006:178–184. doi: 10.1182/asheducation-2006.1.178. [DOI] [PubMed] [Google Scholar]
- 3.Goemans B.F., et al. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005;19:1536–1542. doi: 10.1038/sj.leu.2403870. [DOI] [PubMed] [Google Scholar]
- 4.Streubel B., et al. Amplification of the MLL gene on double minutes, a homogeneously staining region, and ring chromosomes in five patients with acute myeloid leukemia or myelodysplastic syndrome. Genes Chromosomes Cancer. 2000;27:380–386. [PubMed] [Google Scholar]
- 5.Ferrando A.A., et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Fernandez J. Hox, ParaHox, ProtoHox: facts and guesses. Heredity. 2005;94:145–152. doi: 10.1038/sj.hdy.6800621. [DOI] [PubMed] [Google Scholar]
- 7.Pineault N., Helgason C.D., Lawrence H.J., Humphries R.K. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T., et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 9.Schoch C., et al. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102:2395–2402. doi: 10.1182/blood-2003-02-0434. [DOI] [PubMed] [Google Scholar]
- 10.Camos M., et al. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–6954. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- 11.Kim D.Y., et al. Upregulated hoxC4 induces CD14 expression during the differentiation of acute promyelocytic leukemia cells. Leuk. Lymphoma. 2005;46:1061–1066. doi: 10.1080/10428190500102589. [DOI] [PubMed] [Google Scholar]
- 12.Scholl C., et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. . J. Clin. Invest. 2007;117:1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James R., Kazenwadel J. Homeobox gene expression in the intestinal epithelium of adult mice. J. Biol. Chem. 1991;266:3246–3251. [PubMed] [Google Scholar]
- 14.Chase A., et al. Fusion of ETV6 to the caudal-related homeobox gene CDX2 in acute myeloid leukemia with the t(12;13)(p13;q12). Blood. 1999;93:1025–1031. [PubMed] [Google Scholar]
- 15.Rawat V.P., et al. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 2004;101:817–822. doi: 10.1073/pnas.0305555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giampaolo A., et al. Expression pattern of HOXB6 homeobox gene in myelomonocytic differentiation and acute myeloid leukemia. Leukemia. 2002;16:1293–1301. doi: 10.1038/sj.leu.2402532. [DOI] [PubMed] [Google Scholar]
- 17.Bansal D., et al. Cdx4 dysregulates Hox gene expression and generates acute myeloid leukemia alone and in cooperation with Meis1a in a murine model. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16924–16929. doi: 10.1073/pnas.0604579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partanen J., Schwartz L., Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev. 1998;12:2332–2344. doi: 10.1101/gad.12.15.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roche J., et al. Hox expression in AML identifies a distinct subset of patients with intermediate cytogenetics. Leukemia. 2004;18:1059–1063. doi: 10.1038/sj.leu.2403366. [DOI] [PubMed] [Google Scholar]
- 20.Muller-Tidow C., et al. High-throughput analysis of genome-wide receptor tyrosine kinase expression in human cancers identifies potential novel drug targets. Clin. Cancer Res. 2004;10:1241–1249. doi: 10.1158/1078-0432.ccr-0954-03. [DOI] [PubMed] [Google Scholar]
- 21.Veraksa A., Del Campo M., McGinnis W. Developmental patterning genes and their conserved functions: from model organisms to humans. Mol. Genet. Metab. 2000;60:85–100. doi: 10.1006/mgme.2000.2963. [DOI] [PubMed] [Google Scholar]