Abstract
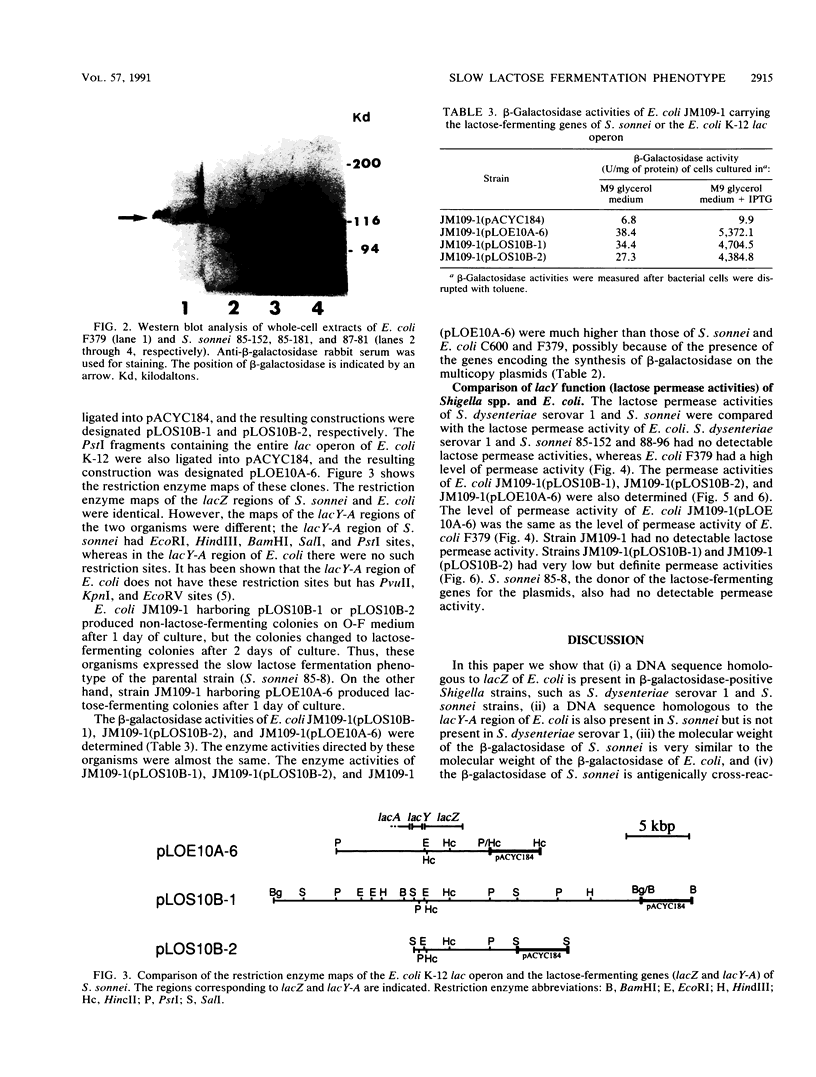
A Southern hybridization analysis revealed that the region homologous to Escherichia coli lacZ was present on the chromosomal DNAs of beta-galactosidase-positive Shigella strains, such as Shigella dysenteriae serovar 1 and Shigella sonnei strains, whereas this region was absent from chromosomal DNAs of beta-galactosidase-negative strains of Shigella flexneri and Shigella boydii. We found that the lacY-A region was deficient in S. dysenteriae serovar 1 and believe that this is the reason for the slow fermentation of lactose by this strain. S. sonnei strains possessed the region which hybridized with E. coli lacY-A despite their slow hydrolysis of lactose. The whole lactose-fermenting region was cloned from S. sonnei and compared with the cloned lac operon of E. coli K-12. Both clones directed the synthesis of beta-galactosidase in an E. coli K-12 strain lacking indigenous beta-galactosidase activity (strain JM109-1), and we observed no difference in the expression of beta-galactosidase activity in S. sonnei and E. coli. However, E. coli JM109-1 harboring the lactose-fermenting genes of S. sonnei exhibited the slow lactose fermentation phenotype like the parental strain. S. sonnei strains had no detectable lactose permease activities. E. coli JM109-1 harboring the lactose-fermenting genes of S. sonnei had a detectable permease activity, possibly because of the multicopy nature of the cloned genes, but this permease activity was much lower than that of strain JM109-1 harboring the lac operon of E. coli K-12. From these results we concluded that slow lactose fermentation by S. sonnei is due to weak lactose permease activity.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arakawa Y., Ohta M., Kido N., Mori M., Ito H., Komatsu T., Fujii Y., Kato N. Chromosomal beta-lactamase of Klebsiella oxytoca, a new class A enzyme that hydrolyzes broad-spectrum beta-lactam antibiotics. Antimicrob Agents Chemother. 1989 Jan;33(1):63–70. doi: 10.1128/aac.33.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R. J., Myster S. H., Wilson T. H. Characterization and sequencing of the lac Y54-41 "uncoupled" mutant of the lactose permease. J Biol Chem. 1989 May 15;264(14):8135–8140. [PubMed] [Google Scholar]
- Kido N., Ohta M., Iida K., Hasegawa T., Ito H., Arakawa Y., Komatsu T., Kato N. Partial deletion of the cloned rfb gene of Escherichia coli O9 results in synthesis of a new O-antigenic lipopolysaccharide. J Bacteriol. 1989 Jul;171(7):3629–3633. doi: 10.1128/jb.171.7.3629-3633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Stauffer G. V., Plamann M. D., Stauffer L. T. Construction and expression of hybrid plasmids containing the Escherichia coli glyA genes. Gene. 1981 Jun-Jul;14(1-2):63–72. doi: 10.1016/0378-1119(81)90148-7. [DOI] [PubMed] [Google Scholar]