Abstract
The heterochromatin protein 1 (HP1) family is thought to be an important structural component of heterochromatin. HP1 proteins bind via their chromodomain to nucleosomes methylated at lysine 9 of histone H3 (H3K9me). To investigate the role of HP1 in maintaining heterochromatin structure, we used a dominant negative approach by expressing truncated HP1α or HP1β proteins lacking a functional chromodomain. Expression of these truncated HP1 proteins individually or in combination resulted in a strong reduction of the accumulation of HP1α, HP1β, and HP1γ in pericentromeric heterochromatin domains in mouse 3T3 fibroblasts. The expression levels of HP1 did not change. The apparent displacement of HP1α, HP1β, and HP1γ from pericentromeric heterochromatin did not result in visible changes in the structure of pericentromeric heterochromatin domains, as visualized by DAPI staining and immunofluorescent labeling of H3K9me. Our results show that the accumulation of HP1α, HP1β, and HP1γ at pericentromeric heterochromatin domains is not required to maintain DAPI-stained pericentromeric heterochromatin domains and the methylated state of histone H3 at lysine 9 in such heterochromatin domains.
INTRODUCTION
The HP1 (heterochromatin protein 1) gene was first discovered in Drosophila melanogaster as a mutant suppressing position effect variegation, a phenomenon in which a gene adjacent to a heterochromatin domain shows a mosaic expression pattern (James and Elgin, 1986; Eissenberg et al., 1990; Festenstein et al., 1999). Since then, a variety of HP1 homologues has been found in many eukaryotes (Li et al., 2002). There are at least three HP1 proteins in mammals. HP1α and HP1β are present in heterochromatic regions, whereas HP1γ has been found either exclusively in euchromatin or in both euchromatin and heterochromatin (Wreggett et al., 1994; Horsley et al., 1996; Minc et al., 1999, 2000; Cheutin et al., 2003). HP1 proteins are thought to have a role in gene regulation in a broad range of eukaryotes: from fission yeast to mammals (Hiragami and Festenstein, 2005; Hediger and Gasser, 2006).
The HP1 gene products are proteins of ∼180 amino acids, containing two protein-binding domains linked by a flexible hinge region. The chromodomain (CD) located at the N-terminal side of the hinge region specifically binds histone H3 methylated at lysine 9 (H3K9me; Bannister et al., 2001; Lachner et al., 2001; Jacobs and Khorasanizadeh, 2002), which is a marker for heterochromatin and is related to gene silencing (Cowell et al., 2002; Sims et al., 2003). HP1 is able to repress gene activity when artificially targeted to a site near a reporter gene (van der Vlag et al., 2000). The C-terminal part of HP1 contains the chromoshadow domain (CSD), which is partially homologous to the CD (Aasland and Stewart, 1995; Sims et al., 2003). The CSD binds to a consensus peptide that is present in a number of proteins, including the CSD of HP1 itself, thereby targeting proteins to heterochromatin and forming HP1 dimers (Brasher et al., 2000; Smothers and Henikoff, 2000; Nielsen et al., 2001; Li et al., 2002; Thiru et al., 2004). Finally, HP1 has been shown to bind RNA, probably at its hinge region. RNA binding is important for binding the protein to pericentromeric heterochromatin (Muchardt et al., 2002).
HP1 proteins are assumed to have a structural role in defining the condensed heterochromatin state. HP1 dimers may bridge nucleosomes in heterochromatin via H3K9me, resulting in a compact chromatin structure (Jenuwein, 2001; Nielsen et al., 2001; Lachner and Jenuwein, 2002; Singh and Georgatos, 2002). However, there is no direct evidence that HP1 is an essential component of heterochromatin (Singh and Georgatos, 2002). In mouse fibroblasts, pericentromeric heterochromatin constitutes conspicuous nuclear domains that can be visualized by DAPI staining. HP1 proteins are highly accumulated in these domains. A large fraction of the HP1 in heterochromatin domains rapidly exchanges with nucleoplasmic HP1, whereas a small fraction exchanges slowly, as shown by FRAP and FCS studies (Cheutin et al., 2003; Festenstein et al., 2003; Schmiedeberg et al., 2004).
In this study we investigate whether HP1 accumulation is essential for maintaining pericentromeric heterochromatin domains that can be visualized by DAPI staining in mouse fibroblasts. Using a dominant negative approach in which truncated forms of HP1α and HP1β that lack a functional chromodomain are overexpressed, we show that the apparent displacement of HP1α, HP1β, and HP1γ from pericentromeric heterochromatin does not result in visible changes in large-scale chromatin structure of DAPI-stained heterochromatin domains and in H3K9me levels of such domains. Results indicate that accumulation of HP1α, HP1β, or HP1γ at pericentromeric heterochromatin is not required to maintain DAPI-stained pericentromeric heterochromatin domains and the methylated state of histone H3 at lysine 9 in such heterochromatin domains.
MATERIALS AND METHODS
DNA Constructs
HP1α and HP1β denote the full-length sequences of the human HP1s. HP1α-Δ(2-39) and HP1β-Δ(2-40) have deletions of amino acids 2-39 and 2-40 in HP1α and HP1β, respectively. These proteins lack a large part of the chromodomain, including V21 (HP1α) and V23 (HP1β), which are required for MeH3K9 binding (Platero et al., 1995; Bannister et al., 2001; Lachner et al., 2001). HP1α, HP1β, HP1α-Δ(2-39), and HP1β-Δ(2-40) fragments were PCR-amplified and cloned into a tetracycline-inducible vector (pUHD10-3; Gossen and Bujard, 1992) at the N-terminal end of an inserted flag-tag. The truncated HP1β-Δ(2-40) DNA fragment was digested with EcoRI and BamHI and inserted into pmCherry-C1 (Shaner et al., 2004), resulting in pmCherry-HP1β-Δ(2-40). EGFP-tagged HP1β was kindly provided by Drs. L. Schmiedeberg and P. Hemmerich (Schmiedeberg et al., 2004).
Cell Culture and Transfection
NIH/3T3 mouse fibroblasts were cultured in DMEM supplied with 10% tetracycline-free fetal bovine serum (BD Biosciences, Clontech, San Jose, CA) in a 5% CO2 atmosphere. Cells were grown on coverslips coated with Alcian blue 8GS (Fluka, Buchs, Switzerland). Cells were transfected using FuGENE-6 (Roche, Basel, Switzerland) by adding 0.5 μg DNA of each plasmid and 6 μl of FuGene-6 reagent per milliliter culture medium. Fibroblasts were cotransfected with pUHD15-1, containing the tTA gene, necessary for the Tet-ON system (Gossen and Bujard, 1992). We used pEGFP-C1 (BD Biosciences-Clontech) for expressing enhanced green fluorescent protein (eGFP). NIH/3T3 cells stably expressing EGFP-tagged HP1β were selected using 800 μg/ml G418. A low expressing cell population was sorted by FACS analysis. Cells expressing EGFP-HP1β were transiently transfected with pmCherry-HP1β-Δ(2-40) and analyzed between 24 and 48 h after transfection.
Immunolabeling and DAPI Staining
Cells growing on coverslips were washed twice with PBS and fixed with 2% formaldehyde for 15 min at room temperature (RT) and permeabilized with 0.5% (wt/vol) Triton X-100 in PBS for 5 min. Residual aldehyde groups were blocked for 10 min with 0.1 M glycine PBS at RT. Incubation with primary antibodies in PBG (0.5% (wt/vol) BSA, 0.1% (wt/vol) gelatin in PBS) was performed for 2 h at RT and with the secondary antibodies for 1 h in the same buffer at RT. DAPI staining was carried out by incubation for 5 min in 0.4 μg/ml DAPI in PBS.
Antibodies
To detect flag-tagged proteins we used rabbit anti-flag and mouse anti-flag M2 (Sigma-Aldrich; Zwijndrecht, The Netherlands). To detect HP1α mouse-anti-HP1α (MaHP1α; Euromedex; Mundolsheim, France; Nielsen et al., 1999) and rabbit-anti-HP1α (RaHP1α; Kourmouli et al., 2000) were used. For detection of HP1β we used mouse-anti-HP1β (MaHP1β; Euromedex; Nielsen et al., 1999) and rat-anti-HP1β (RaaHP1β; Wreggett et al., 1994). We used mouse anti-HP1γ (MaHP1γ; Euromedex; Nielsen et al., 1999) to detect HP1γ. RaHP1α and RaaHP1β were kindly provided by Dr. P. B. Singh. All anti-HP1 antibodies recognize human as well as murine HP1. For in situ labeling of H3K9me we used rabbit anti-trimethylated H3K9 (RaH3K9me3; Cowell et al., 2002), also provided by Dr. P. B. Singh; rabbit anti-dimethylated H3K9 (RaH3K9me2; Upstate, Milton Keynes, United Kingdom; Nakayama et al., 2001), and rabbit anti-branched (4×) methylated peptide-H3K9 (RaH3K9me-branched; Maison et al., 2002) provided by Dr. T. Jenuwein. As secondary antibodies we used donkey anti-rabbit IgG(H+L)-Cy5, donkey anti-rabbit IgG(H+L)-Cy3, donkey anti-mouse IgG(H+L)-Cy3, donkey anti-mouse IgG(H+L)-FITC, donkey anti-rabbit IgG(H+L)-FITC, and donkey anti-rat IgG-FITC (Jackson ImmunoResearch Laboratories, West Grove, PA).
Microscopy and Image Analysis
Images were recorded with a Zeiss LSM 510 confocal laser scanning microscope equipped with C-Apochromat 63×/1.2 water immersion lenses (Carl Zeiss Jena GmbH, Jena, Germany). All tracks were recorded separately to reduce cross-talk. Three or more midnuclear confocal sections (0.5–1 μm apart) were recorded for every image. We made line scans in the x-y plane. Figures were composed in Adobe PhotoShop v.7.0 (Adobe Systems, San Jose, CA). For quantitative image analysis, maximum intensity projections of the imaged cells (represented in the Supplementary Figure S1) were generated. Average signal intensity was measured for every channel over circular areas of 1.5 μm2. These circles were placed over five DAPI-dense chromatin regions and five non-DAPI-dense chromatin regions in transfected and nontransfected control cells. The ratio between transfected (dn) and nontransfected cells (wt), for the DAPI signal, as well as for the endogenous HP1 signal, was calculated in individual images as the ratio between the differences between average intensity in DAPI-dense heterochromatic chromatin regions (HC) and non-DAPI-dense euchromatic regions (EC) according to the following: (HC − EC)dn/(HC − EC)wt.
To quantify the levels of EGFP-tagged HP1β in living cells before and after pmCherry-HP1β-Δ(2-40) expression, we imaged the cells on a Zeiss Axiovert 200M widefield fluorescence microscope, equipped with a 100× Plan-Apochromat (1.4 NA) oil immersion lens (Zeiss) and a Cairn Xenon Arc lamp with monochromator (Cairn Research, Kent, United Kingdom). The objective was temperature-controlled with an objective heater, and cells were examined in microscopy medium (137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 20 mM d-glucose, and 20 mM HEPES) at 37°C. Images were recorded with a cooled CCD camera (Coolsnap HQ, Roper Scientific, Tucson, AZ). A 375–490 excitation filter, 490 long-pass dichroic mirror, and 525–40 band-pass emission filter was used for EGFP imaging (monochromator: 470 ± 20 nm). A 375–580 excitation filter, 585 long-pass dichroic mirror, and 620–60 band-pass emission filter was used for mCherry imaging (monochromator: 550 ± 20 nm). The average pixel intensity in the nucleus of the imaged EGFP-HP1β–expressing cells was quantified using Metamorph software (Molecular Devices, Sunnyvale, CA). The distribution of pixel intensities (ranging from 0 to 1600) of 50 cells of each population (transfected and nontransfected) was plotted in a histogram (32 bins with a width of 50).
RESULTS
Approach
We used a dominant negative approach to analyze the role of HP1 in maintaining pericentromeric heterochromatin in the interphase mouse nucleus. Human HP1α and HP1β were cloned as full-length FLAG-tagged proteins (HP1α-flag and HP1β-flag) and as FLAG-tagged truncated proteins (HP1α-Δ(2-39)-flag and HP1β-Δ(2-40)-flag) that lack most of the CD, which is essential for binding to H3K9me (Platero et al., 1995; Bannister et al., 2001; Lachner et al., 2001). To identify transfected cells, a FLAG-tag was positioned at the C-terminal end of all constructs. We used 3T3 mouse fibroblasts, which have large pericentromeric heterochromatin domains that are easily visible after DAPI staining. Cells were cotransfected with the tetracycline-inducible vector pUHD15–1 and the wild-type or mutant HP1 construct in the absence of tetracycline or doxycycline. Expression of all transfected proteins (full-length and truncated HP1) was detectable 12 h after transfection using an anti-flag antibody (data not shown). After 24 h, cells with high expression levels were analyzed for HP1 localization and DAPI-stained heterochromatin domains.
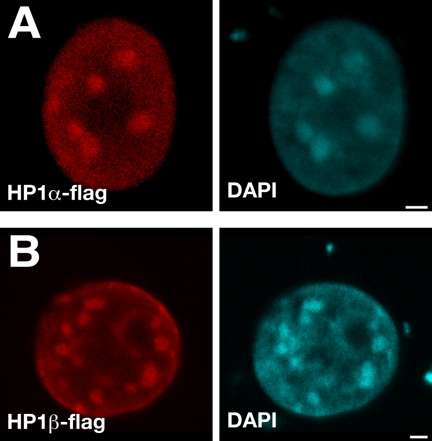
Immunofluorescent labeling, using mouse-anti-flag antibodies (Figure 1) and rabbit-anti-flag (data not shown), showed that the transfected wild-type HP1α-flag and HP1β-flag proteins are targeted to the DAPI-intense stained heterochromatin domains in the nucleus, as shown also by others (Wreggett et al., 1994; Horsley et al., 1996; Minc et al., 1999; Cheutin et al., 2003). The same anti-flag antibodies showed that HP1α-Δ(2-39)-flag and HP1β-Δ(2-40)-flag proteins are localized in the nucleus. As expected, HP1α-Δ(2-39)-flag and HP1β-Δ(2-40)-flag did not accumulate in the DAPI-intense areas, because they lack the CD (Figure 2, red and cyan channels). In most of these cells the anti-flag signal is homogenously distributed in the nucleus, whereas in some cells the anti-flag signal is more concentrated at the periphery of the cell nucleus. Our results agree with the notion that the CD is required for localization of HP1α and HP1β in DAPI-stained pericentromeric heterochromatin through H3K9me binding (Bannister et al., 2001; Lachner et al., 2001; Cowell et al., 2002; Jacobs and Khorasanizadeh, 2002).
Figure 1.
Distribution of transfected full-length HP1α-flag and HP1β-flag in mouse fibroblasts. Cells transfected with HP1α-flag (A) and HP1β-flag (B) were fluorescently immunolabeled 24 h after transfection. The red signal (mouse anti-flag) shows the distribution of transfected HP1α-flag (A) and HP1β-flag (B). The cyan channel shows the DAPI staining. Bars, 2 μm. Individual midnuclear optical sections are shown.
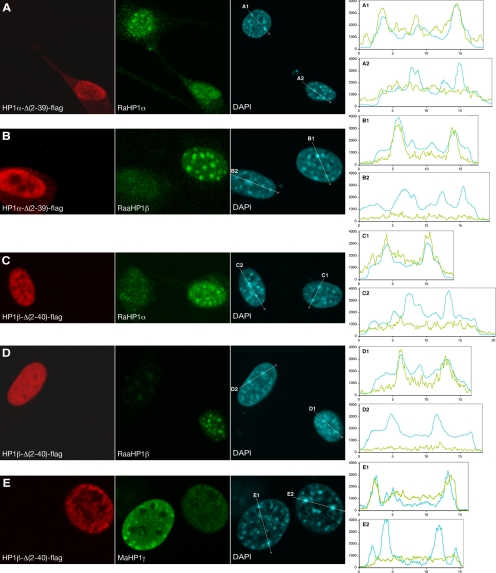
Figure 2.
Spatial distribution of transfected truncated HP1, endogenous HP1, and DAPI-stained heterochromatin domains. Cells transfected with HP1α-Δ(2-39)-flag (A and B) and HP1β-Δ(2-40)-flag (C–E) were fluorescently labeled 24 h after transfection. The red signal (Maflag in A–D and Raflag in E) shows the transfected HP1α-Δ(2-39)-flag (A and B) and HP1β-Δ(2-40)-flag (C–E). The green signal shows the distribution of endogenous HP1α, HP1β, and HP1γ after labeling with RaHP1α (A and C), RaaHP1β (B and D), and MaHP1γ (E). The cyan signal shows the DAPI staining. On the right, intensity profiles are shown of endogenous HP1α, HP1β, and HP1γ; green line) and DAPI stain (cyan line) along the lines shown in the DAPI images. A1, B1, C1, D1, and E1 are line scans in control cells, and A2, B2, C2, D2, and E2 in transfected cells. The x-axis shows distance in μm; the y-axis represents signal intensity in arbitrary units. Individual midnuclear optical sections are shown.
Truncated HP1α and HP1β Displace Endogenous HP1α, HP1β, and HP1γ from Pericentromeric Heterochromatin
Endogenous HP1α, HP1β, and HP1γ in nontransfected cells are located in the nucleus and accumulate in DAPI intense pericentromeric heterochromatin domains (Figure 2 and the second and third column of the Supplementary Figures 1, A–I; Wreggett et al., 1994; Horsley et al., 1996; Minc et al., 1999, 2000; Cheutin et al., 2003). To establish whether truncated HP1 displaces the endogenous protein from heterochromatin domains, cells were transfected with HP1α-Δ(2-39)-flag (Figure 2, A and B, and Supplementary Figure S1, A–D), or HP1β-Δ(2-40)-flag (Figure 2, C–E, and Supplementary Figure S1, E–I). After 24 h, cells were fixed and fluorescently labeled with antibodies against flag-tagged transfected proteins (Figure 2, red channel, and Supplementary Figure S1, first column) and with antibodies against HP1α, HP1β, or HP1γ (Figure 2, green channel, and Supplementary Figure S1, second column) and stained with DAPI (Figure 2, cyan channel, and Supplementary Figure S1, third column). For detection of HP1α we used two different antibodies, i.e., a mouse-anti-HP1α mAb (MaHP1α; Supplementary Figure S1, B and F) and a rabbit anti-HP1α polyclonal antibody (RaHP1α; Figure 2, A and C, and Supplementary Figure S1, A and E). The two antibodies gave identical results. Cells expressing HP1α-Δ(2-39)-flag showed a strong increase in labeling intensity by MaHP1α, but not by RaHP1α labeling. This indicates that MaHP1α recognizes the transfected HP1α-Δ(2-39)-flag protein, whereas RaHP1α does not. For detection of endogenous HP1β we used a mouse anti-HP1β mAb (MaHP1β; Supplementary Figure S1, D and H) and a rat-anti-HP1β mAb (RaaHP1β; Figure 2, B and D, and Supplementary Figure S1, C and G). Both anti-HP1β antibodies did not recognize the mutant HP1β-Δ(2-40)-flag protein. To detect HP1γ, we used a monoclonal mouse-anti-HP1γ (MaHP1γ; Figure 2E and Supplementary Figure S1I). The specificity of all antibodies has been tested elsewhere (see references in Materials and Methods). As a control we analyzed nontransfected cells in the same preparation to relate HP1 accumulation at pericentromeric heterochromatin in control cells to the amount of HP1 in these domains in transfected cells. To allow semiquantitative assessment of the colocalization and intensity of the different labels, line scans were made in the x-y plane through intense DAPI domains in transfected (Figure 2, A2, B2, C2, D2, and E2) and in nontransfected control cells (Figure 2, A1, B1, C1, D1, and E1).
In cells transfected with HP1α-Δ(2-39)-flag, HP1α labeling with RaHP1α showed a dispersed distribution of the HP1α antigen throughout the nucleus, lacking accumulation in the DAPI-stained pericentromeric regions (Figure 2A and Supplementary Figure S1A). Labeling with MaHP1α also showed no accumulation of HP1α in DAPI-intense domains in HP1α-Δ(2-39)-flag–transfected cells, compared with nontransfected control cells (Supplementary Figure S1B). This indicates that endogenous HP1α is displaced from the DAPI-stained pericentromeric heterochromatin domains by the truncated protein. Strikingly, in HP1α-Δ(2-39)-flag–transfected cells, HP1β labeling with RaaHP1β (Figure 2B and Supplementary Figure S1C) or MaHP1β (Supplementary Figure S1D) also showed no accumulation of endogenous HP1β in DAPI-stained chromocenters. These results show that truncated HP1α displaces endogenous HP1α and HP1β from DAPI-stained pericentromeric heterochromatin.
In HP1β-Δ(2-40)–flag transfected cells, HP1β did not accumulate in the DAPI-stained pericentromeric domains and was dispersed throughout the nucleoplasm, as shown by labeling with two different antibodies, i.e., RaaHP1β (Figure 2D and Supplementary Figure S1G) and MaHP1β (Supplementary Figure S1H). Moreover, in HP1β-Δ(2-40)–flag transfected cells RaHP1α (Figure 2C and Supplementary Figure S1E), MaHP1α (Supplementary Figure S1F) and MaHP1γ (Figure 2E and Supplementary Figure S1I) labeling did not show any accumulation of HP1α and HP1γ in DAPI-stained pericentromeric domains. To verify that the displacement of endogenous HP1 is not a consequence of the transfection procedure, fibroblasts were transfected with a plasmid containing only the enhanced eGFP gene and were subsequently immunofluorescently labeled with MaHP1α and MaHP1β. No changes in the distribution or intensity of endogenous HP1 were observed, compared with nontransfected cells (data not shown). Therefore, it is unlikely that the observed changes in HP1 distribution after expression of the mutant HP1 proteins are an artifact of the transfection procedure.
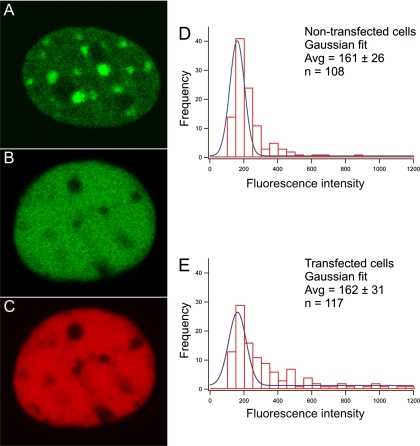
In addition to the analysis of endogenous HP1 levels in fixed cells using anti-HP1 antibodies, we analyzed the dominant negative effect of transfecting cells with truncated HP1 in living cells. To this end we created a mouse fibroblast cell line stably expressing EGFP-tagged HP1β. In these EGFP-HP1β–expressing cells, HP1β was found to accumulate in DAPI-stained pericentromeric heterochromatin domains and to occur diffusely throughout the nucleoplasm (Figure 3A). Transfection of these EGFP-HP1β–expressing cells with mCherry-tagged HP1β-Δ(2-40), resulted in a homogeneous nuclear distribution of both the truncated and the wild-type HP1β proteins (Figure 3, B and C). These results confirm that the antibody labeling data in Figures 2 and 5 and Supplementary Figure S1, showing the truncated HP1 protein that lacks a functional CD displaces full-length HP1 from the DAPI-dense nuclear domains.
Figure 3.
Spatial distribution of EGFP-tagged HP1β-expressing cells transfected with mCherry-tagged HP1β-Δ(2-40). Cells stably expressing EGFP-tagged HP1β show that HP1 is accumulating in pericentromeric heterochromatin domains (A). EGFP-tagged HP1β expressing cells were transfected with mCherry-tagged HP1β-Δ(2-40) (B and C). The green signal shows the EGFP-tagged HP1β (B). The red signal shows the transfected mCherry-tagged HP1β-Δ(2-40) (C). The fluorescence intensity of EGFP-HP1β in cells trasfected with mCherry-tagged HP1β-Δ(2-40) was compared with nontransfected cells. A histogram of the Gaussian distribution of the EGFP-HP1β fluorescence intensity in transfected (E) and nontransfected cells (D) is shown. x-axis, the average fluorescence intensity in arbitrary units; y-axis, the number of cells.
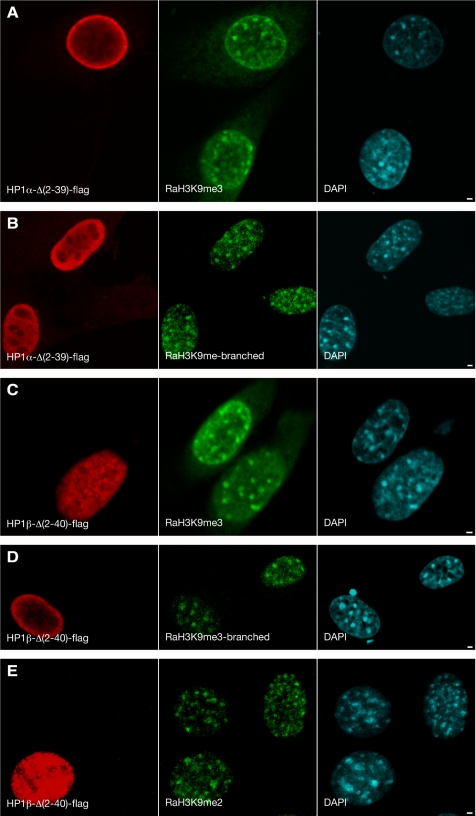
Figure 5.
Spatial distribution of H3K9me in cells transfected with truncated HP1α and HP1β. Cells transfected with HP1α-Δ(2-39)-flag (A and B) and HP1β-Δ(2-40)-flag (C–E) were fluorescently labeled 24 h after transfection. The red signal (mouse-anti-flag) shows the transfected HP1α-Δ(2-39)-flag (A and B) and HP1β-Δ(2-40)-flag (C–E). The green signal shows distribution of H3K9me detected by either RaMeH3K9me3 (A and C), RaH3K9mebranched (B and D), or RaH3K9me2 (D). The cyan signal represents DAPI staining. Bars, 2 μm. Individual midnuclear optical sections are shown.
To examine whether expression of truncated HP1β-Δ(2-40) results in a decrease of the HP1β concentration, we determined the expression level of cells stably expressing EGFP-HP1β with and without transfection with mCherry-HP1β-Δ(2-40) (Figure 3). The fluorescence intensity of EGFP-HP1β integrated over the nucleus was determined 24 h after transfection in cells expressing high levels of mCherry-HP1β-Δ(2-40) (based on the red signal) and in untransfected cells. The distribution of EGFP-HP1β fluorescence measured in transfected and nontransfected cells was fitted with a Gaussian distribution yielding average EGFP-HP1β fluorescence intensities of 161 ± 26 and 162 ± 40 for nontransfected and transfected cells, respectively. These results show that expression of truncated HP1β does not change the cellular concentration of full-length EGFP-HP1β (Figure 3, D and E).
These results show that truncated HP1α and truncated HP1β protein, which lack a functional CD, not only expel their full-length HP1 counterpart from the DAPI-stained pericentromeric heterochromatin domains, but also the two other HP1 homologues. This suggests that the three HP1 homologues interact in heterochromatin.
Loss of HP1α and HP1β from Heterochromatin Does Not Result in Changes in Large-Scale Heterochromatin Structure
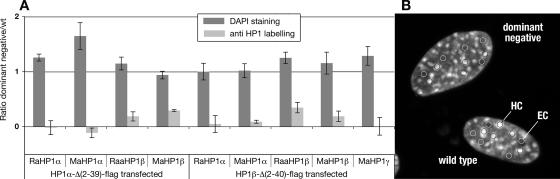
We performed a quantitative analysis of the relative amount of DAPI staining and endogenous HP1 levels in cells transfected with HP1α-Δ(2-39)-flag or HP1β-Δ(2-40)-flag and in untransfected control cells. Results demonstrate that there is no change in DAPI staining after transfection with either HP1α-Δ(2-39)-flag or HP1β-Δ(2-40)-flag. The ratio of DAPI stain in transfected versus nontransfected cells is close to unity (Figure 4). In contrast, the ratio for the HP1 signal, using different antibodies (i.e., RaHP1α, MaHP1α, RaaHP1β, MaHP1β, and MaHP1γ) is close to zero (Figure 4). These measurements show in a quantitative manner that, after expression of HP1 that lacks the chromodomain, most of the endogenous HP1 is relocated from DAPI dense chromatin regions to the nucleoplasm, whereas the DAPI staining remains unchanged.
Figure 4.
Quantitative analysis of the spatial distribution of transfected truncated HP1, endogenous HP1, and DAPI-stained heterochromatin domains. Maximum intensity projections of the imaged cells (see Supplementary Figure S1) were generated. Average signal intensity was measured for every channel over circular areas of 1.5 μm2. These circles were placed over five DAPI dense chromatin regions and five non-DAPI dense chromatin regions in transfected and nontransfected control cells (B). The ratio between cells transfected with HP1α-Δ(2-39)-flag or HP1β-Δ(2-40)-flag versus not transfected control cells, for the DAPI signal (black bars) as well as for the endogenous HP1 signal (gray bars) was calculated. This was done in individual images as the ratio of the differences between average intensity in DAPI dense chromatin regions (HC) and non-DAPI dense euchromatic chromatin regions (EC) in transfected (dn) and nontransfected cells (wt): (HC − EC)dn/(HC − EC)wt (A). Error bars, SE between the different images.
To analyze whether the loss of HP1α and HP1β from DAPI-stained pericentromeric heterochromatin domains is correlated with a decrease in H3K9me level, we analyzed the distribution of H3K9me 24 h after transfection with HP1α-Δ(2-39)-flag, HP1β-Δ(2-40)-flag, or with both simultaneously. We labeled H3K9me with three different antibodies: RaH3K9me2, RaH3K9me3, and RaH3K9me-branched. The RaH3K9me-branched antibody is described to have a high affinity for H3K9me3, to significantly cross-react with di- and trimethylation of several other H3 lysine positions, and to decorate pericentromeric heterochromatin and the inactive X chromosome (Maison et al., 2002; Perez-Burgos et al., 2004; Figure 5, B and D). No detectable change in H3K9me labeling was observed with the RaH3K9me3 and RaH3K9me-branched antibodies 24 h after transfection with either HP1α-Δ(2-39)-flag (Figure 5, A and B), HP1β-Δ(2-40)-flag (Figure 5, C and D), or with HP1α-Δ(2-39)-flag and HP1β-Δ(2-40)-flag simultaneously (data not shown). Similarly, no change in the dimethylation level of H3K9 was observed (Figure 5E). The same results were obtained 38 h after transfection, using RaH3K9me2, RaH3K9me3, and RaH3K9me-branched antibodies (data not shown). These results show that, although all three endogenous HP1 proteins are to a large degree displaced from DAPI-stained pericentromeric heterochromatin domains by HP1α-Δ(2-39)-flag or HP1β-Δ(2-40)-flag, this does not result in a visible reduction of H3K9me levels in these domains.
We showed that the overall structure, size, and nuclear distribution of DAPI-stained pericentromeric heterochromatin domains remained unaffected as the three HP1 proteins are displaced (Figures 2 and 5 and Supplementary Figure S1, A–I). These observations demonstrate that the visible accumulation of HP1α, HP1β, and HP1γ is not essential for maintaining pericentromeric heterochromatin structure as visualized by DAPI staining and by immunofluorescent labeling of H3K9me2, H3K9me3, or H3K9me-branched.
DISCUSSION
HP1 is thought to play an important role in heterochromatin organization and the control of gene expression (Hiragami and Festenstein, 2005; Hediger and Gasser, 2006). This involves the specific binding of the CD to H3K9me, dimerization via its CSD and recruitment of a variety of proteins that specifically interact via HP1 with H3K9me-rich chromatin domains (Brasher et al., 2000; Jenuwein, 2001; Nielsen et al., 2001; Cowell et al., 2002; Lachner and Jenuwein, 2002; Singh and Georgatos, 2002; Thiru et al., 2004). HP1 dimerizes through its CSD and may bridge two H3K9me-containing nucleosomes, inducing a higher order packing of heterochromatin (Jenuwein, 2001; Nielsen et al., 2001; Lachner and Jenuwein, 2002; Singh and Georgatos, 2002; Thiru et al., 2004). It has also been proposed that HP1 dimers bind to methylated K9 and K27 on a single histone H3 tail, instead of binding to methylated K9 on nearby nucleosomes (Jacobs and Khorasanizadeh, 2002). HP1 may also play a role in the maintenance of the heterochromatin state through cell division, via its interactions with chromatin assembly factor 1 (CAF1), H3K9 methyltransferase Suv39, and the DNA methyltransferases Dnmt1 and Dnmt3a (reviewed in Maison and Almouzni, 2004). In this study we used a dominant negative approach to interfere with HP1 function to obtain insight into the role of HP1α, HP1β, and HP1γ to maintain DAPI-stained pericentromeric heterochromatin domains in mouse fibroblasts.
Expressing truncated HP1α and HP1β, lacking a functional CD, resulted in displacement of the endogenous HP1α, HP1β, and HP1γ from the visible DAPI-stained pericentromeric heterochromatin domains (Figure 2). It did not result in a significant decrease in full-length HP1 expression (Figure 3). Remarkably, loss of HP1 did not result in a visible change in the structure of pericentromeric heterochromatin domains, as visualized by DAPI staining, and did not result in a visible change the methylation state of H3K9 (Figures 2, 4, 5 and Supplementary Figure 5). It is known that the CD is responsible for specific binding of HP1 to H3K9me in heterochromatin (Bannister et al., 2001; Lachner et al., 2001; Cowell et al., 2002; Jacobs and Khorasanizadeh, 2002; Cheutin et al., 2003). Therefore, as expected, HP1 lacking a CD did not accumulate in DAPI-stained heterochromatin domains (Figure 2 and Supplementary Figure S1), in contrast to wild-type HP1α-flag and HP1β-flag, which did accumulate in these domains (Figure 1). Evidently, interaction of the CD of the endogenous HP1 with H3K9me is not sufficient to keep the proteins associated with DAPI-stained heterochromatin domains. Most likely, the truncated polypeptides sequester endogenous proteins that are necessary to stabilize the interaction of HP1 with H3K9me. Because expression of the each of the truncated protein individually resulted in the apparent displacement of HP1α, HP1β, and HP1γ from DAPI-stained pericentromeric heterochromatin, it is likely that these three HP1 proteins interact directly or indirectly in vivo.
Our results are consistent with studies showing that the CD is necessary but not sufficient for HP1 to bind H3K9me-containing nucleosomes (Smothers and Henikoff, 2001; Thiru et al., 2004) and that HP1β as a monomeric protein is not stably associated with pericentromeric heterochromatin (Thiru et al., 2004). Our data show that when most endogenous HP1 is displaced from DAPI-dense heterochromatin, the level and distribution of tri- and dimethylated H3K9me remains unchanged. This demonstrates that accumulation of HP1α, HP1β, and HP1γ is not required for keeping histone H3K9 in its methylated state in DAPI-stained heterochromatin domains. In this context, labeling with the H3K9me-branched antibody, as well as HP1α accumulation at pericentromeric heterochromatin, have been found to be lost after incubation with RNAse and after culturing in the presence of a histone deacetylase inhibitor (Maison et al., 2002; Muchardt et al., 2002). Besides, Gilbert et al. (2003) reported that during differentiation of chicken erythrocytes HP1 is completely lost, whereas there is only a modest reduction in H3K9me. Obviously, maintaining H3K9me levels does not require the continuous presence of HP1 proteins.
Interestingly, Peters et al. (2001) presented evidence that DAPI-dense domains, in which HP1 accumulates, remain present in cells that fully lack the histone methyltransferases Suv39h1 and 2. In these cells heterochromatin domains do not contain H3K9me. From these and our results it can be concluded that HP1 binding and H3K9me are independent processes and are not required simultaneously for heterochromatin stability. Recently, we showed that in vivo targeting of HP1α or HP1β to an amplified chromosomal region causes local chromatin condensation, enhanced trimethylated H3K9me, and recruitment of histone methyltransferases (Verschure et al., 2005). Targeting of the HP1 lacking a functional CD also caused heterochromatinization of the amplified chromosome region (Brink et al., 2006). These results show that normal binding of HP1 through its CD domain, as well as artificial binding of HP1 without a CD through lac operator-lac Repressor interaction, is sufficient to trigger heterochromatin formation. The present studies indicate that HP1 is not required for maintaining the DAPI-stained heterochromatin domains.
HP1 in heterochromatin is remarkably dynamic and exchanges rapidly with its soluble pool (Cheutin et al., 2003; Festenstein et al., 2003; Schmiedeberg et al., 2004). HP1 is bound in heterochromatin domains in at least two modes: one that exchanges rapidly and one that is released slowly. The ratio between these pools ranges from ∼8:1 in NIH/3T3 fibroblasts (Schmiedeberg et al., 2004) to close to 3:1 in mouse T-cells (Festenstein et al., 2003), probably depending on cell type and chromatin state. The function of the fast- and the slow-exchanging HP1 subfractions is unknown. Our results show in fixed cells as well as in living cells that in mouse fibroblasts most HP1 protein can be removed from DAPI-stained pericentromeric heterochromatin without a visible change in large-scale chromatin structure or a significant decrease in H3K9me. It is possible that a small fraction of the strongly bound fraction of HP1 molecules, e.g., the slowly exchanging fraction, remains present in the dominant negative experiments. Light microscopic analysis is not sufficiently sensitive to decide this unambiguously. However, our data clearly show that most HP1 in visible pericentromeric domains is dispensable for maintaining typical heterochromatin features, including strong, DAPI staining, H3K9 methylation, and the heterochromatin size and shape. Our findings indicate that local accumulation of HP1α, HP1β, and HP1γ is not essential for the maintenance of DAPI-stained pericentromeric heterochromatin.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. P. B. Singh and to T. Jenuwein for providing us with antibodies used in this study; Dr. R. Y. Tsien (San Diego, CA) for providing the mCherry cDNA; and Drs. L. Schmiedeberg and P. Hemmerich (Jena, Germany) for providing EGFP-tagged HP1β. We thank Dr. E.M.M. Manders and particularly W. Takkenberg, who tragically died in a motorcycle accident, for assistance with confocal microscopy. We also thank Ir. B. Hooibrink, from the Academic Medical Center in Amsterdam for cell sorting. This work was supported in part by the Netherlands Organization for Scientific Research, Earth and Life Sciences (ALW), by an ALW-PULS and ALW-VIDI grant to P.J.V. (project numbers PULS/33-98/805-4811 and VIDI 2003/03921/ALW/016.041.311).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0025) on February 21, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Aasland R., Stewart A. F. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–3174. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Brasher S. V., Smith B. O., Fogh R. H., Nietlispach D., Thiru A., Nielsen P. R., Broadhurst R. W., Ball L. J., Murzina N. V., Laue E. D. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink M. C., van der Velden Y., de Leeuw W., Mateos-Langerak J., Belmont A. S., van Driel R., Verschure P. J. Truncated HP1 lacking a functional chromodomain induces heterochromatinization upon in vivo targeting. Histochem. Cell Biol. 2006;125:53–61. doi: 10.1007/s00418-005-0088-7. [DOI] [PubMed] [Google Scholar]
- Cheutin T., McNairn A. J., Jenuwein T., Gilbert D. M., Singh P. B., Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Cowell I. G., et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., Elgin S. C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festenstein R., Pagakis S. N., Hiragami K., Lyon D., Verreault A., Sekkali B., Kioussis D. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- Festenstein R., Sharghi-Namini S., Fox M., Roderick K., Tolaini M., Norton T., Saveliev A., Kioussis D., Singh P. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat. Genet. 1999;23:457–461. doi: 10.1038/70579. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Boyle S., Sutherland H., de Las Heras J., Allan J., Jenuwein T., Bickmore W. A. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 2003;22:5540–5550. doi: 10.1093/emboj/cdg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Gasser S. M. Heterochromatin protein 1, don't judge the book by its cover! Curr. Opin. Develop. 2006;16:143–150. doi: 10.1016/j.gde.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Hiragami K., Festenstein R. Heterochromatin protein 1, a pervasive controlling influence. Cell Mol. Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley D., Hutchings A., Butcher G. W., Singh P. B. M32, a murine homologue of Drosophila heterochromatin protein 1 (HP1), localises to euchromatin within interphase nuclei and is largely excluded from constitutive heterochromatin. Cytogenet. Cell Genet. 1996;73:308–311. doi: 10.1159/000134363. [DOI] [PubMed] [Google Scholar]
- Jacobs S. A., Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- James T. C., Elgin S. C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Kourmouli N., Theodoropoulos P. A., Dialynas G., Bakou A., Politou A. S., Cowell I. G., Singh P. B., Georgatos S. D. Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J. 2000;19:6558–6568. doi: 10.1093/emboj/19.23.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Li Y., Kirschmann D. A., Wallrath L. L. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA. 2002;99(suppl 4):16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004;5:296–305. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A. H., Quivy J. P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Minc E., Allory Y., Worman H. J., Courvalin J. C., Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- Minc E., Courvalin J. C., Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Guilleme M., Seeler J. S., Trouche D., Dejean A., Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen A. L., Ortiz J. A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A. L., Oulad-Abdelghani M., Ortiz J. A., Remboutsika E., Chambon P., Losson R. Heterochromatin formation in mammalian cells. Interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- Perez-Burgos L., Peters A.H., Opravil S., Kauer M., Mechtler K., Jenuwein T. Generation and characterization of methyl-lysine histone antibodies. Methods Enzymol. 2004;376:234–254. doi: 10.1016/S0076-6879(03)76016-9. [DOI] [PubMed] [Google Scholar]
- Peters A. H., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Platero J. S., Hartnett T., Eissenberg J. C. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeberg L., Weisshart K., Diekmann S., Meyer Zu Hoerste G., Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol. Biol. Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans M. N., Palmer A. E., Tsien R. Y. Improbed monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;12:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sims R. J., 3rd, Nishioka K., Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Singh P. B., Georgatos S. D. HP1, facts, open questions, and speculation. J. Struct. Biol. 2002;140:10–16. doi: 10.1016/s1047-8477(02)00536-1. [DOI] [PubMed] [Google Scholar]
- Smothers J. F., Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- Smothers J. F., Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiru A., Nietlispach D., Mott H. R., Okuwaki M., Lyon D., Nielsen P. R., Hirshberg M., Verreault A., Murzina N. V., Laue E. D. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J., den Blaauwen J. L., Sewalt R. G., van Driel R., Otte A. P. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J. Biol. Chem. 2000;275:697–704. doi: 10.1074/jbc.275.1.697. [DOI] [PubMed] [Google Scholar]
- Verschure P. J., van der Kraan I., de Leeuw W., van der Vlag J., Carpenter A. E., Belmont A. S., van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol. Cell. Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreggett K. A., Hill F., James P. S., Hutchings A., Butcher G. W., Singh P. B. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.