Abstract
The minichromosome maintenance genes (MCM2-7) are transcribed at M/G1 just as the Mcm complex is imported into the nucleus to be assembled into prereplication complexes, during a period of low cyclin-dependent kinase (CDK) activity. The CDKs trigger DNA replication and prevent rereplication in part by exporting Mcm2-7 from the nucleus during S phase. We have found that repression of MCM2-7 transcription in a single cell cycle interferes with the nuclear import of Mcms in the subsequent M/G1 phase. This suggests that nascent Mcm proteins are preferentially imported into the nucleus. Consistent with this, we find that loss of CDK activity in G2/M is not sufficient for nuclear import, there is also a requirement for new protein synthesis. This requirement is not met by constitutive production of Cdc6 and does not involve synthesis of new transport machinery. The Mcm proteins generated in the previous cell cycle, which are unable to reaccumulate in the nucleus, are predominantly turned over by ubiquitin-mediated proteolysis in late mitosis/early G1. Therefore, the nuclear localization of Mcm2-7 is dependent on nascent transcription and translation of Mcm2-7 and the elimination of CDK activity which occurs simultaneously as cells enter G1.
INTRODUCTION
Within each cell cycle, the entire genome must be replicated exactly once. This complex process involves the assembly and activation of dozens of proteins, a process that is controlled by the periodic activity of the cyclin-dependent kinases (CDKs). During early G1 when CDK activity is low, the prereplication complex (pre-RC) is assembled at origins of DNA replication (for review, see Bell and Dutta, 2002). The origin recognition complex (ORC) binds to the origins throughout the cell cycle and serves as the foundation for pre-RC assembly. ORC recruits both Cdc6 and Cdt1 to the origin, and they, in turn, load the hexameric minichromosome maintenance complex (Mcm2-7) to complete the assembly of the pre-RC. DNA replication is triggered by the Cdc7/Dbf4 kinase and the B-type CDKs. The B-type CDKs also inhibit any new pre-RC assembly until the next G1, thereby preventing catastrophic rereplication (Broek et al., 1991; Dahmann et al., 1995).
All of the pre-RC components are negatively regulated by the CDKs to prevent origins from firing more than once. The mechanism of CDK regulation has been worked out in most detail in the budding yeast Saccharomyces cerevisiae. CDKs inactivate ORC by phosphorylating two of its six subunits and directly interacting with Orc6 (Nguyen et al., 2001; Wilmes et al., 2004). Cdc6 is targeted for degradation in a CDK-dependent manner (Elsasser et al., 1999; Drury et al., 2000). CDK activity triggers the export of Cdt1 (Tanaka and Diffley, 2002) and soluble Mcm proteins from the nucleus (Labib et al., 1999; Nguyen et al., 2000). If all of these mechanisms are disabled, a modest level of rereplication can be detected (Nguyen et al., 2001; Wilmes et al., 2004; Green et al., 2006; Tanny et al., 2006).
The only pre-RC components known to function beyond the initiation of DNA replication are the Mcm proteins (Labib et al., 2000). This hexameric complex moves with the fork during DNA replication, functioning as the putative DNA helicase (Aparicio et al., 1997; Ishimi, 1997; You et al., 1999; Lee and Hurwitz, 2000; Kaplan et al., 2003). The Mcm proteins are tightly regulated to prevent any inappropriate DNA replication. The nuclear localization of the Mcm2-7 complex is regulated by the CDKs. Mcm2-7 are imported into the nucleus when CDK activity is low in early G1 and exported from the nucleus during S phase when CDK activity is high (Labib et al., 1999; Nguyen et al., 2000). The high CDK activity through G2/M keeps the Mcm proteins from accumulating in the nucleus until the next G1 when pre-RCs are reformed. The Mcm complex contains both a single bipartite nuclear localization signal (NLS) and a nuclear export signal (NES) (Liku et al., 2005). The NLS is split between Mcm2 and Mcm3 and the NES is located in Mcm3 adjacent to the NLS sequence. The transport of all six Mcm proteins is interdependent, suggesting that a hexameric complex must first be formed for nuclear import, which would result in the assembly of a complete NLS (Nguyen et al., 2000; Labib et al., 2001). Phosphorylation of the CDK sites around the NES is required for nuclear exit during S phase, presumably as Mcm2-7 proteins disassociate from the DNA (Liku et al., 2005).
The MCM2-7 genes are also periodically transcribed with the peak of transcription occurring at the end of mitosis (Hennessy et al., 1990; Dalton and Whitbread, 1995; McInerny et al., 1997; Pramila et al., 2002) when Mcm2-7 begin to concentrate in the nucleus. All six MCM genes contain an early cell cycle box (ECB) transcriptional element in their promoter (McInerny et al., 1997; Pramila et al., 2002). It is constitutively bound by the transcriptional activator Mcm1 and periodically inhibited by the homeodomain proteins Yhp1 and Yox1. Either Yox1 or Yhp1 is present from late G1 through early mitosis, restricting the transcription of the MCM2-7 genes to late mitosis and early G1.
The temporal correlation between MCM2-7 transcription and nuclear entry led us to determine the importance of the transcriptional regulation of the MCM genes in the assembly of the pre-RC. This was done by tracking the nuclear localization of the Mcm complex, which is a prerequisite for the assembly of the pre-RC. We found that lowering CDK activity was not sufficient to recycle Mcms into the nucleus. At least two of the proteins (Mcm3 and Mcm4) were largely degraded before the next G1, and nascent protein synthesis was required along with lowered CDK activity to enable Mcms to accumulate in the nucleus. Moreover, preventing MCM2-7 transcription in a single cell cycle significantly impeded their nuclear localization. We conclude that nascent transcription and translation of the Mcms in M/G1 and the degradation of cytoplasmic Mcm proteins generated in the previous cycle both contribute to the efficient nuclear localization of Mcm2-7 protein complexes.
MATERIALS AND METHODS
Strains and Growth Conditions
The strains, plasmids, and oligonucleotides used in this study are listed in Supplemental Tables S1–S3. The MCM4-GFP integrating plasmid (BD2818) was constructed by first inserting a EcoRI–BamHI fragment of green fluorescent protein (GFP) (enhanced GFP with S65T and F64L mutations) into pRS306, followed by the insertion of a C-terminal fragment of MCM4 (171 amino acid) upstream of GFP, which was amplified with KpnI and EcoRI sites and a 12-amino acid linker at the C terminus (GAGAGAGAGAEF). BD2818 was linearized with Tth111I and integrated at the MCM4 locus in BY2125 (WT) and BY3087 (GAL-YOX1) (Pramila et al., 2002) to generate BY3944 (MCM4-GFP) and BY3945 (MCM4-GFP GAL-YOX1). The strains used for the chromosome stability experiments were derived from YPH363, which contains an extra chromosome marked with URA3 and SUP11 (Spencer et al., 1990). YPH363 was transformed with GAL2 and the mating type was changed to MATa by using a GAL-HO plasmid to generate BY4254. YOX1 was placed under the GAL1 promoter in BY4254 to generate BY4265 (GAL-YOX1) as described previously (Pramila et al., 2002). The TEF-CDC6 integrating plasmid (BD2878) was constructed by amplifying CDC6 with BamHI and XhoI sites and inserting into pRS415-TEF and then subcloning the SacI–XhoI fragment of TEF-CDC6 into pRS305. BD2878 was linearized with BglII and integrated at the CDC6 locus of BY3944 and BY3945 to generate BY4485 (TEF-CDC6 MCM4-GFP) and BY4487 (TEF-CDC6 GAL-YOX1 MCM4-GFP). The GAL-CDC14 plasmid (BD2836) was constructed by amplifying CDC14 with HindIII and XhoI sites and inserting into pRS415-GAL1. BY3944 and BY3945 were transformed with BD2836 to generate BY5781 (pGAL-CDC14 MCM4-GFP) and BY5782 (pGAL-CDC14 GAL-YOX1 MCM4-GFP). The KpnI–BamHI fragment of MCM4-GFP from BD2818 was subcloned into pRS304 to generate BD2859, which was linearized with Tth111I and integrated into YKL83 (GAL-UBR1) and YKB7 (cdc28-td GAL-UBR1) (Noton and Diffley, 2000) to generate BY5225 (GAL-UBR1 MCM4-GFP) and BY5226 (cdc28-td GAL-UBR1 MCM4-GFP). The pML104 plasmid (pRS304-GAL1-NLS2-NLS3NES-GFP3) (Liku et al., 2005) was linearized with EcoRV and integrated at the TRP1 locus in BY2125 and BY3087 to generate BY5649 (GAL1-NLS2-NLS3NES-GFP3) and BY5652 (GAL1-NLS2-NLS3NES-GFP3 GAL-YOX1). YAT187 (SWI6-GFP; Geymonat et al., 2004) was transformed with BD2836 to generate BY6066 (pGAL-CDC14 SWI6-GFP). Cells were grown in rich yeast extract and peptone media with 2% glucose, galactose, or raffinose except for BY5781, BY5782, and BY6066, which were grown in minimal synthetic complete (SC) media without leucine to maintain plasmid. Then, 5 μg/ml α-factor was used to arrest cells in G1 and 10 μg/ml nocodazole was used to arrest cells in G2/M; 10 μg/ml cycloheximide was used to inhibit protein synthesis.
Microscopy
To monitor the localization of Mcm4-GFP and Swi6-GFP, we examined either live cells or fixed cells by microscopy. For live cells, 1 ml of cells was collected, washed with PS buffer (0.1 M potassium phosphate, pH 7.5, and 1.2 M sorbitol) and stained with 4′,6-diamidino-2-phenylindole (DAPI) as below. For fixed cells, 1 ml of cells were collected, resuspended in 100 μl of paraformaldehyde, and incubated at room temperature for 15 min. Cells were washed with PS buffer, resuspended in the same buffer with 0.5% Triton X-100 and 1 μg/ml DAPI, and incubated at room temperature for 5 min. These cells were washed with PS buffer and resuspended in a small volume to spot on slides. The prepared cells were examined in a Nikon Eclipse E600 microscope with a Nikon Plan Apochromat 60 × A/1.40 oil immersion objective lens by using either a fluorescein isothiocyanate-HYQ filter (excitation at 460–500) for Mcm4-GFP or a UV-2E/C DAPI filter (excitation at 330–380) for DAPI-stained DNA. Photographs of cells were taken with a Photometrics Cascade 512B camera by using the MetaMorph version 6.3r2 software (Molecular Devices, Sunnyvale, CA). Brightness and contrast were adjusted in Canvas equally for samples in the same experiment. The number of cells was counted that were unbudded or had divided their nuclei as determined by DAPI, and then the localization of Mcm4-GFP was scored.
Flow Cytometry
To monitor progression through the cell cycle, cells stained with Sytox-Green (Invitrogen, Carlsbad, CA) were analyzed by flow cytometry. 0.5 ml of cells were collected for each sample and fixed with 1 ml of ethanol overnight. Cells were washed with H2O and incubated with 0.2 mg/ml RNase A in 50 mM Tris·HCl, pH 8, for 4 h. The cells were collected and incubated with 2 mg/ml proteinase K in 50 mM Tris·HCl, pH 7.5, for 1 h The cells were washed with fluorescence-activated cell sorting (FACS) buffer (200 mM Tris·HCl, pH 7.5, 200 mM NaCl, and 78 mM MgCl2), resuspend in 5 μM Sytox-Green in FACS buffer, and sonicated. Stained cells were analyzed on a FACScan cytometer (BD Biosciences, San Jose, CA).
RNA Measurements
Levels of mRNA were measured using an S1 nuclease protection assay with probes for MCM3 (BL642), MCM4 (BL1408), and ACT1 (BL606) as described previously (Pramila et al., 2002). Sequences of probes are listed in Supplemental Table S3.
Immunoblotting
To analyze the levels of Mcm3 and Mcm4, either equal total protein or an equal number of cells was loaded for each sample. Mcm3, Orc3 and Swi6 were detected with Mcm3-18 (Liang and Stillman, 1997), Orc3-SB3 (Liang and Stillman, 1997), and 4550 (Sidorova and Breeden, 1993), respectively. GFP was detected with A. v. peptide antibody from Clontech (Mountain View, CA), and tubulin was detected with YOL1/34 antibody from Accurate Chemical and Scientific Corporation (Westbury, NY). For chemiluminescence detection, anti-mouse IgG horseradish peroxidase (HRP) and anti-rabbit IgG HRP (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) were used as the secondary antibodies and developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical, Rockford, IL). Alexa Fluor 680 goat anti-mouse IgG and Alexa Fluor 680 goat anti-rat IgG fluorescent secondary antibodies (Invitrogen) were detected with the Odyssey scanner (Li-Cor Biosciences, Lincoln, NE).
Chromosome Stability
BY4254 (WT) and BY4265 (GAL-YOX1), which contain an extra chromosome, were grown in raffinose and plated on SC media with low adenine (6 μg/ml adenine) and glucose to determine background chromosome loss. Then, cells were grown for 18 h in galactose and plated on SC media with low adenine and glucose to determine the chromosome loss after galactose induction. The number of cells was counted at the beginning and end of the induction by using a Z2 Coulter counter (Beckman Coulter, Fullerton, CA) to determine the number of generations that elapsed. The number of red (lost extra chromosome) and white (retained extra chromosome) colonies was determined, and the number of red colonies per generation was calculated, which is equivalent to the number of chromosomes lost per generation (Spencer et al., 1990).
RESULTS
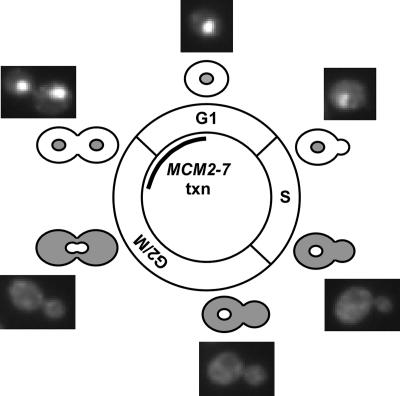
It has been previously shown that the nuclear localization of the Mcm proteins is dependent on all six subunits (Nguyen et al., 2000; Labib et al., 2001); therefore, the localization of all six Mcm proteins can be accessed by examining the localization of a single subunit of the Mcm2-7 complex. To monitor Mcm2-7 localization, we fused a GFP tag to the C terminus of the genomic MCM4 coding sequence (Labib et al., 1999). Figure 1 shows that the bulk of Mcm4 is nuclear in G1 through early S phase when it is exported from the nucleus. At the M/G1 border, Mcm4 distribution changes again and becomes nuclear, just as prereplication complexes are formed. This shift to the nucleus is temporally correlated with the new transcription of all six Mcm proteins. This led us to determine whether the transcriptional regulation of the MCM genes was important for their efficient assembly into the pre-RC.
Figure 1.
Localization of Mcm4-GFP through the cell cycle. The microscopy images of Mcm4-GFP fluorescence are shown at different stages of the cell cycle, and nuclear localization was determined by colocalization with DAPI staining of the DNA in BY3944 (MCM4-GFP). The cartoons of yeast cells show the known localization of Mcm2-7 represented as gray shading (Labib et al., 1999; Nguyen et al., 2000). txn, transcription.
Reduced CDK Activity and Protein Synthesis Is Required before Nuclear Import of the Mcm Complex
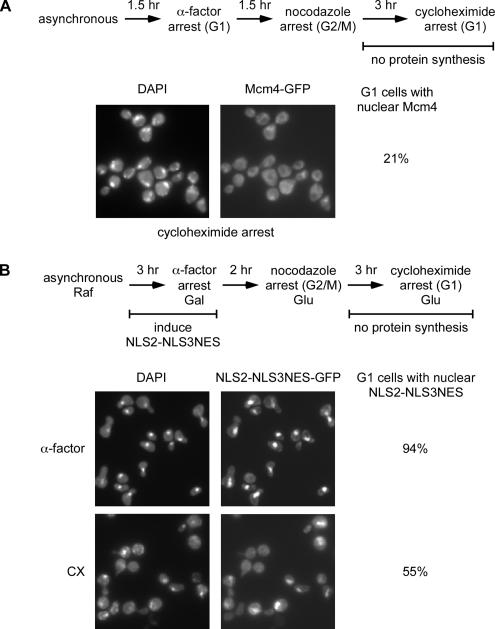
If nascent Mcm proteins preferentially enter the nucleus, we predict that there should be a strong requirement for protein synthesis late in the cycle to achieve their redistribution to the nucleus. To address this issue, we arrested cells in G2/M with nocodazole and then released them into the protein synthesis inhibitor cycloheximide. Figure 2A shows that after 3 h in cycloheximide, most of the cells have exited mitosis and are unbudded G1 cells. However, the Mcm4 signal remains cytoplasmic. The Mcm4 signal is weak and strikingly uniform in these cells, with only 21% showing a slight enrichment in the nucleus.
Figure 2.
Protein synthesis is required for the nuclear accumulation of Mcm4. (A) BY3944 (MCM4-GFP) is arrested in G1 with α-factor, released directly into nocodazole to arrest in G2/M, and then released into cycloheximide to block protein synthesis while progressing to G1. The Mcm4-GFP fluorescence and DAPI-stained DNA images from the G1 arrest in cycloheximide are shown. The percentage of cells with divided nuclei that have nuclear Mcm4-GFP in the cycloheximide-induced G1 arrest is shown to the right of the images. (B) BY5649 (GAL-NLS2-NLS3NES-GFP3) was grown in raffinose and switched to galactose with α-factor to induce NLS2-NLS3NES for 3 h and arrest the cells in G1. Cells were released into nocodazole to obtain a G2/M arrest in the presence of glucose to repress transcription of NLS2-NLS3NES and then released into cycloheximide to inhibit protein synthesis. The microscopy images of NLS2-NLS3NES-GFP fluorescence and DAPI-stained DNA are shown for the first α-factor arrest and the after cycloheximide arrest. The percentage of G1 cells with NLS2-NLS3NES concentrated in the nucleus in both the initial G1 arrest and the second G1 arrest in cycloheximide is quantitated and shown to the right of the images. NLS2-NLS3NES, Mcm transport module; cx, cycloheximide; Raf, raffinose; Gal, galactose; Glu, glucose.
We then followed the localization of an artificial construct referred to as the Mcm transport module in the same experiment. This module is a fusion of three GFP moieties to the NLS sequences from Mcm2 and Mcm3 and the nuclear export signal from Mcm3 (NLS2-NLS3NES-GFP). It has been shown to contain all the necessary localization signals to confer the same cell cycle regulated and CDK-dependent nuclear import and export that is observed with the intact Mcm2-7 complex, but it is transcribed ectopically from the GAL1 promoter (Liku et al., 2005). In contrast to the localization pattern with full-length Mcm4-GFP expressed from its endogenous promoter, this GAL-driven construct localizes to the nucleus in the majority of the cells, despite the cycloheximide treatment (Figure 2B). Cells were initially grown in raffinose and then transferred to galactose in the presence of the yeast pheromone α-factor to induce the Mcm transport module while arresting in G1. These cells were released into nocodazole to induce a G2/M arrest while simultaneously repressing the transcription of the Mcm transport module with glucose. Finally, cells were released from the G2/M block into cycloheximide to inhibit protein synthesis while still allowing progression into G1. The Mcm transport module was concentrated in the nucleus in the first G1 in 94% of the cells (Figure 2B). After cycloheximide treatment (CX), the Mcm transport module synthesized in the first G1 shows a similarly strong nuclear enrichment in 55% of the cells in the second G1. This demonstrates that the transport machinery required for nuclear import of the Mcm2-7 proteins remains active in cycloheximide and can efficiently recycle the Mcm transport module back into the nucleus. This experiment also serves as an important control for the ability of cells arrested with nocodazole and released into cycloheximide to progress to the low CDK, G1-like state required for nuclear transport directed by the Mcm transport signals.
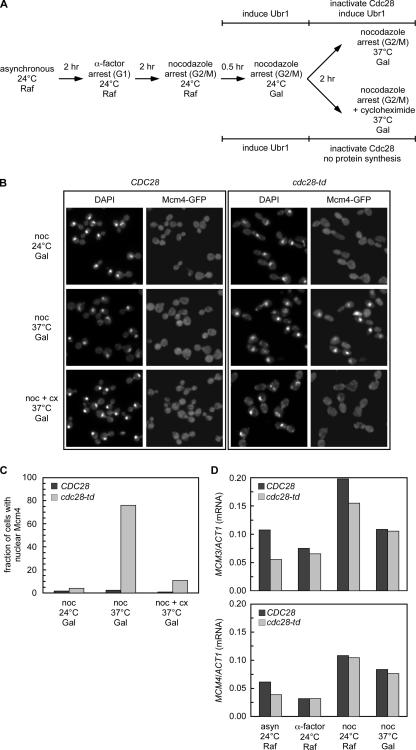
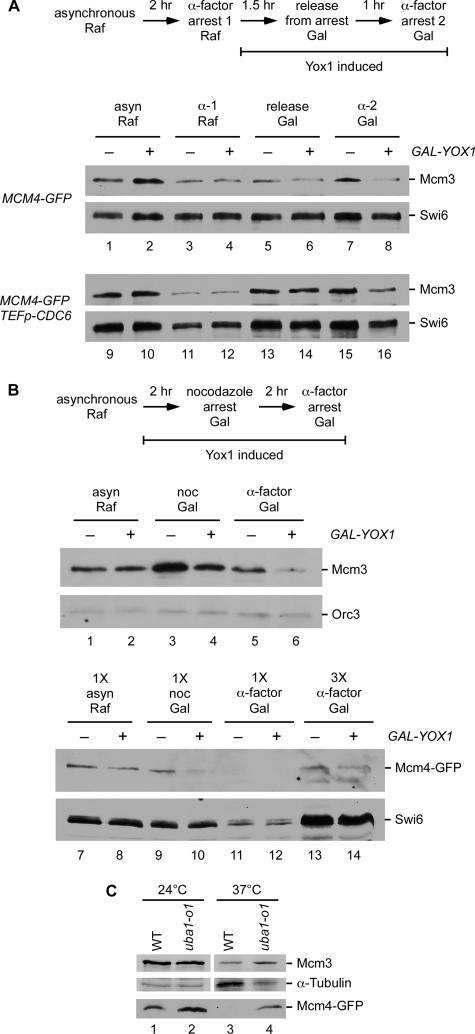
Previous studies have shown that the Mcm2-7 proteins are prevented from accumulating in the nucleus during mitosis because of the high CDK activity. Labib et al. (1999) showed that if you inactivate the CDKs by degrading the Cdc28 kinase with a temperature- and Ubr1-sensitive allele of CDC28 (cdc28-td) during a nocodazole arrest (G2/M), the Mcm2-7 proteins accumulate in the nucleus. This indicates that loss of CDK activity is necessary to enable Mcm2-7 to enter the nucleus. However, Mcms are transcribed during a nocodazole arrest (Figure 3D), so it is possible that only the nascent Mcms are able to enter the nucleus when CDK activity is eliminated. We addressed this question by repeating the cdc28-td experiment in the presence of cycloheximide. cdc28-td cells were synchronized with α-factor and released into nocodazole to arrest in G2/M at the permissive temperature, and then the GAL-UBR1 construct was induced by adding galactose. These cells were then shifted to the nonpermissive temperature to eliminate CDK activity while maintaining the arrest in G2/M (Figure 3A). In the absence of cycloheximide, we see that 76% of the cells have Mcm4 concentrated in the nucleus when Cdc28-td is degraded (Figure 3, B and C), consistent with the published results. However, the addition of cycloheximide blocks this nuclear accumulation of Mcm4. These results suggest that during G2/M, CDK activity prevents both old and newly synthesized Mcms from accumulating in the nucleus. When CDK activity drops late in mitosis, only the newly synthesized Mcm proteins are competent for nuclear import.
Figure 3.
CDK inactivation and protein synthesis are required for nuclear accumulation of Mcm4 in mitosis. BY5225 (GAL-UBR1 MCM4-GFP) and BY5226 (cdc28-td GAL-UBR1 MCM4-GFP) were arrested in α-factor, released into nocodazole to arrest in G2/M at 24°C in raffinose. Galactose was added to induce Ubr1 and then the cells were transferred to 37°C in galactose to induce the degradation of Cdc28-td while maintaining the G2/M arrest. Cycloheximide is added to one set of cultures to inhibit protein synthesis. (A) Experimental outline. (B) The Mcm4-GFP fluorescence and DAPI-stained DNA images for the nocodazole arrest at 24 and 37°C as well as the nocodazole arrest at 37°C with cycloheximide are shown for both strains. (C) Quantitation of microscopy images showing the fraction of cells with Mcm4-GFP concentrated in the nucleus is plotted. (D) Levels of MCM3 and MCM4 mRNA relative to ACT1 mRNA as determined by an S1 nuclease protection assay are plotted for each of the arrests. noc, nocodazole.
Overexpression of Yox1 Inhibits Mcm2-7 Nuclear Localization
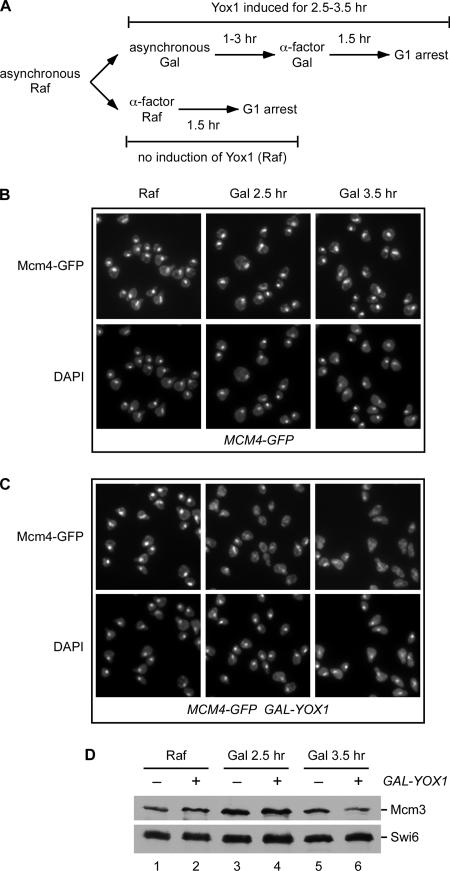
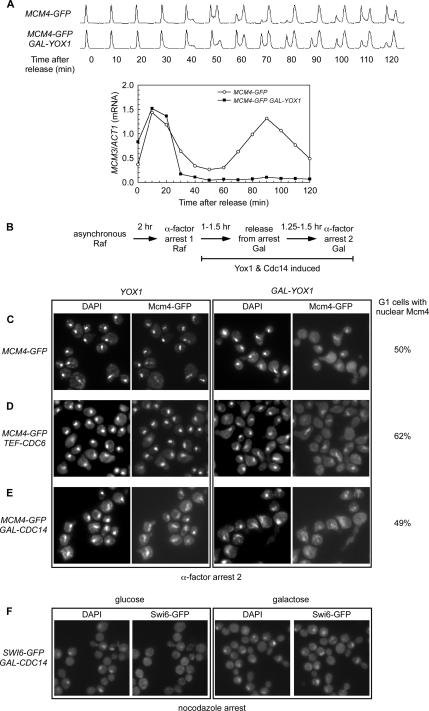
MCM2-7 are periodically transcribed and peak at the M/G1 transition due to their repression by two homeodomain proteins, Yox1 and Yhp1, from S to late M phase. This interval of peak MCM2-7 transcription coincides with the dramatic shift of Mcm2-7 from the cytoplasm to the nucleus (Figure 1). Overexpression of Yox1 represses transcription of MCM2-7, causes a severe growth defect and leads to an S-phase delay (Pramila et al., 2002). These observations led us to investigate the role of nascent transcription in the nuclear localization of Mcm2-7. To see whether MCM transcription impacts this pattern of localization, we looked at the localization of Mcm4 when Yox1 was overexpressed. We placed YOX1 under the GAL1 promoter, which allowed us to greatly overexpress Yox1 when cells were grown in galactose. Previous experiments have shown that GAL-YOX1 causes a 3- to 30-fold drop in the levels of MCM mRNAs (Pramila et al., 2002). Cells were initially grown in raffinose, and then they were switched to galactose to induce Yox1 for 2.5 and 3.5 h (Figure 4A). α-factor was added to arrest these cells in G1 and Mcm4-GFP localization was monitored. In the absence of Yox1 induction (Figure 4, B and C, Raf), all the cells in both the control strain and the GAL-YOX1 strain concentrated Mcm4 in the nucleus in the G1 arrest. However, the overexpression of Yox1 for 2.5 and 3.5 h resulted in a dramatic loss of Mcm4 nuclear accumulation (Figure 4C). To ensure that Mcm protein was present during the course of the galactose induction, Mcm3 levels were assessed at each time point (Figure 4D). Mcm3 protein levels were equal at 2.5 h and only slightly reduced after 3.5 h. This suggests that Yox1 interferes with the localization of the Mcm2-7 complex either directly by inhibiting MCM2-7 transcription or indirectly by inhibiting the transcription of a factor required for Mcm2-7 nuclear transport.
Figure 4.
Overexpression of Yox1 disrupts Mcm4 nuclear accumulation. BY3944 (MCM4-GFP) and BY3945 (GAL-YOX1 MCM4-GFP) were grown in raffinose, and then galactose was added to induce Yox1. α-Factor was added for the last 1.5 h of the galactose incubation. The localization of Mcm4-GFP was monitored by fluorescence and compared with DAPI-stained DNA. This was compared with the cells that were grown and arrested with α-factor in raffinose. (A) Experimental outline. (B and C) The microscopy images are shown for cells arrested in the second G1 with α-factor. (D) Mcm3 protein levels were monitored during the experiment. Equal amounts of total protein were loaded, and Swi6 protein served as a loading control.
The reduced nuclear pool of Mcm proteins in the GAL-YOX1 strain would be expected to lead to increased chromosome loss and an extended S phase (Lei et al., 1996; Liang et al., 1999; Fitch et al., 2003; Liku et al., 2005). Consistent with this, the GAL-YOX1 strain has an extended S phase as shown in the FACS in Figure 5A (Pramila et al., 2002). In addition, we find that the GAL-YOX1 strain containing a marked extra chromosome has a chromosome loss rate of 19% per generation compared with only 0.4% per generation for the WT strain (see Materials and Methods for details). This 50-fold increase in chromosome loss due to the overexpression of Yox1 shows that regulation of Yox1 expression is important in maintaining chromosome stability.
Figure 5.
Overexpression of Yox1 disrupts Mcm4 nuclear accumulation in a single cycle. (A) BY3944 (MCM4-GFP) and BY3945 (GAL-YOX1 MCM4-GFP) were grown in raffinose, arrested in α-factor, and released into galactose to induce Yox1. Cells were collected every 10 min during the induction. DNA content was determined by flow cytometry. RNA was isolated from cells at each time point and the levels of MCM3 mRNA compared with ACT1 mRNA were determined using an S1 nuclease protection assay and plotted. (B) Experimental outline for C–E. (C–E) BY3944 (MCM4-GFP), BY3945 (GAL-YOX1 MCM4-GFP), BY4485 (TEF-CDC6 MCM4-GFP), BY4487 (GAL-YOX1 TEF-CDC6 MCM4-GFP), BY5781 (GAL-CDC14 MCM4-GFP), and BY5782 (GAL-YOX1 GAL-CDC14 MCM4-GFP) were grown in raffinose, arrested in α-factor, and released into galactose to induce Yox1 and Cdc14. Mcm4-GFP localization was examined in the next G1 by arresting the cells in α-factor and Mcm4-GFP fluorescence and DAPI-stained DNA are shown in the microscopy images. The percentage of cells without buds or with divided nuclei were scored for Mcm4-GFP nuclear accumulation for the second α-factor arrest in the GAL-YOX1 strains and shown to the right of the images. (F) BY6066 (SWI6-GFP GAL-CDC14) was grown in raffinose, arrested in nocodazole and then released into either glucose (repress GAL-CDC14) or galactose (induce GAL-CDC14) for 2 h. Swi6-GFP fluorescence and DAPI-stained DNA are shown after the final incubation in the microscopy images.
Transcription of Mcm2-7 Directly Preceding Their Nuclear Localization Is Important
The peak of MCM2-7 transcription directly precedes the accumulation of the Mcm2-7 complex in the nucleus. Therefore, we wanted to determine whether this burst of transcription was required for Mcm2-7 nuclear localization. We used the GAL-YOX1 strain to repress the transcription of the MCM2-7 genes. To ensure that overexpression of Yox1 could repress MCM2-7 transcription in a single cycle, the levels of MCM3 mRNA were assayed over the course of the experiment. Cells grown in raffinose were arrested in α-factor and released into galactose to induce Yox1. As expected, MCM3 mRNA levels peak in α-factor and again at the M/G1 boundary in the control strain (Figure 5A, open circles), but when Yox1 is overexpressed the wave of transcription preceding the second G1 is completely repressed (Figure 5A, closed squares). This allowed us to examine the effect of the loss of transcription at M/G1 on the localization of the Mcm2-7 complex in the subsequent G1. The localization of Mcm4 was examined in the next G1 by arresting the cells again with α-factor (Figure 5B). We found that there was a twofold decrease in cells with Mcm4 concentrated in the nucleus and those cells that did have Mcm4 in the nucleus had a significantly lower level of fluorescence than the control cells (Figure 5C). These results indicate that the M/G1 pulse of transcription that is repressed by Yox1 is important for the proper localization of the Mcm complex.
CDC6 transcription is also repressed by Yox1 (Pramila et al., 2002), and it is required for loading Mcm2-7 into the pre-RCs (Aparicio et al., 1997; Tanaka et al., 1997). To see whether CDC6 repression was responsible for this defect in Mcm localization, we repeated the experiment with a strain expressing CDC6 constitutively from the TEF promoter. Figure 5D shows that there is little or no improvement, with about half the cells showing weak nuclear Mcm4 in the second G1. Therefore, CDC6 repression by Yox1 is not responsible for the defect in Mcm localization. This is consistent with previous results which showed that Cdc6 is not required for nuclear accumulation of Mcm2-7 when CDK activity is low in early G1 (Labib et al., 1999; Nguyen et al., 2000).
Another group of genes inhibited by Yox1 encode proteins involved in late mitosis. These include members of the FEAR and MEN pathways (Pramila et al., 2002). These pathways release and activate the phosphatase Cdc14, which is known to remove phosphates from CDK substrates and promote their nuclear entry (for review, see Stegmeier and Amon, 2004). To see whether Cdc14 is limiting under these conditions, we simultaneously overexpressed both Yox1 and Cdc14 in a single cell cycle experiment as outlined in Figure 5B. Once again, we saw the same defect in Mcm4 localization as with cells overexpressing Yox1 alone (Figure 5, C and E). Previous studies have shown that the induction of a GAL-CDC14 construct during a nocodazole arrest (G2/M) is sufficient to drive efficient nuclear import of another CDK target, Swi6 (Geymonat et al., 2004). We repeated this experiment using the GAL-CDC14 construct we generated and got the same results as previously published (Figure 5F). This indicates that our GAL-CDC14 construct is expressed and active. However, we see no indication that excess Cdc14 can drive Mcm nuclear localization. We conclude that Mcms differ from other proteins exported from the nucleus by CDK phosphorylation in that excess ectopic Cdc14 is not sufficient to drive them back into the nucleus.
Yox1 Does Not Repress the Transcription of Other Factors Needed for Mcm2-7 Nuclear Import
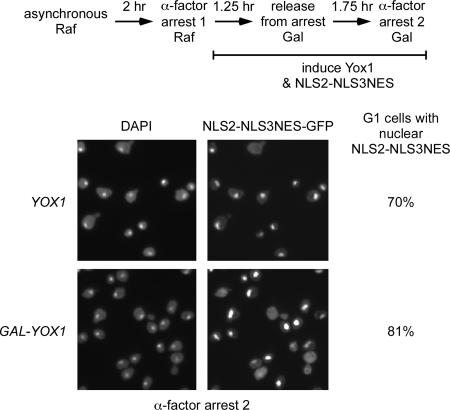
There may be additional proteins required for Mcm nuclear import that must be newly synthesized in mitosis before Mcm nuclear accumulation. To test this, we looked at the localization of the Mcm transport module upon Yox1 overproduction, which interferes with nuclear accumulation of the Mcm2-7 complex (Figures 4 and 5). The cells were arrested with α-factor in raffinose and then released into galactose to induce both the Mcm transport module and Yox1, which are both regulated by the GAL1 promoter. These cells were allowed to proceed into the next G1. In contrast to the effect observed with full-length Mcm protein, Figure 6 shows that overexpression of Yox1 has no impact on the nuclear accumulation of the Mcm transport module in G1. We conclude that the transport machinery that act upon the Mcm nuclear import and export signals are not inhibited directly or indirectly by Yox1 overexpression.
Figure 6.
Nuclear accumulation of the Mcm transport module is independent of Yox1 and protein synthesis. BY5649 (GAL-NLS2-NLS3NES-GFP3) and BY5652 (GAL-YOX1 GAL-NLS2-NLS3NES-GFP3) were grown in raffinose, arrested in α-factor, and then released into galactose to induce Yox1 and NLS2-NLS3NES. The microscopy images of NLS2-NLS3NES-GFP fluorescence and DAPI-stained DNA are shown for the second α-factor arrest. The percentage of G1 cells with NLS2-NLS3NES concentrated in the nucleus in the second G1 is shown to the right of the images.
Old Mcm Protein Is Degraded as Cells Exit Mitosis
If the old Mcms do not return to the nucleus, then we expected that they must be turned over in the cytoplasm to give rise to the dramatic shift in localization that is observed (Figure 1). To test this, we used GAL-YOX1 to inhibit a single cycle of MCM2-7 transcription and examined what happens to the old Mcm protein. As before, cells grown in raffinose were arrested in G1 with α-factor, released into galactose to induce Yox1 and then arrested again in α-factor. We found that the level of Mcm3 protein was much lower in the second G1 arrest in the GAL-YOX strain relative to the control strain (Figure 7A, lanes 7 and 8). This was also the case for the strain constitutively expressing Cdc6 (lanes 15 and 16). A similar result is seen with cells transiting from a nocodazole arrest to G1. The Mcm3 protein present in the G2/M arrest (Figure 7B, lanes 3 and 4) was significantly reduced in the subsequent G1 when transcription was repressed (lane 6). We also looked at the levels of Mcm4-GFP and the results were similar. The levels of Mcm4-GFP dropped off as cells progressed from the G2/M arrest to G1. In both experiments, the majority of the Mcm protein was degraded as cells transit to the next G1 in the absence of new Mcm2-7 synthesis. Mcm3 and Mcm4 seem to be targeted for degradation, but in wild-type growing cells this degradation is balanced by nascent synthesis, leading to the impression that the pool of Mcms remains fairly constant (Hennessy et al., 1990; Dalton and Whitbread, 1995; Young and Tye, 1997).
Figure 7.
Mcm3 and Mcm4 protein are degraded during G2/M. (A) The experimental outline is shown at the top. BY3944 (MCM4-GFP), BY3945 (GAL-YOX1 MCM4-GFP), BY4485 (TEF-CDC6 MCM4-GFP), and BY4487 (GAL-YOX1 TEF-CDC6 MCM4-GFP) were grown in raffinose (lanes 1, 2, 9, and 10), arrested in G1 with α-factor (lanes 3, 4, 11, and 12), and then released into galactose to induce Yox1 (lanes 5, 6, 13, and 14 are 90 min after release) followed by a second α-factor arrest (lanes 7, 8, 15, and 16). Mcm3 protein levels were monitored during the experiment. Equal amounts of total protein were loaded and Swi6 protein serves as a loading control. (B) The experimental outline is shown at the top. BY3944 and BY3945 were grown in raffinose (lanes 1, 2, 7, and 8), arrested in G2/M with nocodazole in galactose to induce Yox1 (lanes 3, 4, 9, and 10), and then released into α-factor to arrest in G1 (lanes 5, 6, 11, 12, 13, and 14). Lanes 13 and 14 have 3 times (3X) as much protein as lanes 7–12. Equal amounts of total protein were loaded and the levels of Mcm3 and Mcm4-GFP protein were monitored over the course of the experiment. Orc3 and Swi6 serve as loading controls. (C) Mcm3 and Mcm4-GFP protein levels were compared in BY5466 (WT) and BY5467 (uba1-o1) at 24°C (permissive temperature) and 37°C (nonpermissive temperature). Equal numbers of cells were loaded, and α-tubulin served as a loading control. Samples were run on the same gel, but a longer exposure was required to visualize Mcm3 at 37°C. asyn, asynchronous; α-1, first α-factor arrest; α-2, second α-factor arrest; noc, nocodazole arrest; α-factor, α-factor arrest.
The degradation of the Mcms could be due to a ubiquitin-mediated mechanism because Mcm3 has been shown to be specifically ubiquitinated in mitosis, and this ubiquitinated form disappears as cells enter G1 (Cheng et al., 2002). To test this, we looked at the protein levels of Mcm3 and Mcm4-GFP in a strain with a temperature-sensitive allele of Uba1 (uba1-o1), which is the only E1 or ubiquitin-activating enzyme in S. cerevisiae (McGrath et al., 1991; Shimada et al., 2002). At the permissive temperature, the levels of Mcm3 and Mcm4-GFP were similar in both the control and uba1-o1 strain, but at the restrictive temperature the levels of Mcm3 and Mcm4-GFP protein were much higher in the E1-deficient strain (Figure 7C). This supports the idea that Mcm3 and Mcm4 are turned over by a ubiquitin-mediated mechanism.
DISCUSSION
Nuclear localization of the Mcm2-7 proteins is an important and highly regulated process. Mcm2-7 are key components of the pre-RC and serve as a putative helicase during DNA synthesis. This complex of six proteins must be synthesized, assembled, and imported into the nucleus to participate in DNA replication. However, this process must be tightly regulated to prevent reassembly of the pre-RC and potential rereplication. Previous studies have shown the importance of CDK activity in promoting nuclear export of the Mcms and the requirement for low CDK activity to promote nuclear import (Labib et al., 1999; Nguyen et al., 2000). These observations have led to a model where CDK-dependent phosphorylation restricts Mcms to the cytoplasm and then this modification is reversed by a phosphatase, which enables the Mcms to reenter the nucleus. One important fact that is not considered in this model is that the MCM2-7 genes are coordinately transcribed and peak at the M/G1 boundary, precisely when the Mcms are imported into the nucleus. Therefore, we set out to test whether elimination of CDK activity was the only requirement for Mcm nuclear accumulation or whether the periodic transcription of the MCMs was also important.
If new transcription of MCM2-7 is required before nuclear transport, then elimination of CDK activity should not be sufficient to relocalize the old Mcms. This was tested by using a Cdc28 with a temperature-sensitive degron, which was degraded during a G2/M arrest when Mcms are cytoplasmic. The degradation of Cdc28-td led to the accumulation of Mcms in the nucleus as described previously, but we found that there was also a requirement for protein synthesis. This was also seen in experiments where we blocked protein synthesis as cells progress from mitosis to G1. This was not due to loss of the Mcm transport machinery, because the Mcm transport module containing all the signals for Mcm nuclear export and import shows strong nuclear accumulation in G1 in the absence of protein synthesis. This is consistent with previous results that showed that both the inactivation of CDK activity and protein synthesis are required for the assembly of pre-RCs (Dahmann et al., 1995; Noton and Diffley, 2000). However, this requirement for protein synthesis was thought to be due only to the need for synthesis of Cdc6. We have directly tested that by including a constitutive source of Cdc6 and find that nuclear localization of Mcms is still defective in that strain. We conclude that nascent Cdc6 and Mcm complex components are required with each cycle of pre-RC formation. Interestingly, CDC6 and MCM2-7 are coordinately transcribed by ECB elements in their promoters (McInerny et al., 1997; Pramila et al., 2002).
The transcription of MCM2-7 is restricted to M/G1 by the transcription factors Yox1 and Yhp1. It has been previously shown that all six MCM genes can be repressed by overexpressing one of these homeodomain proteins, Yox1. Interestingly, we found that overexpression of Yox1 interfered with the nuclear accumulation of Mcms, which suggested that their nascent transcription was important for their nuclear localization. By synchronizing the cells, we were able to specifically repress the burst of MCM transcription that precedes Mcm nuclear localization and look at the fate of the Mcms synthesized in the previous cell cycle. We found that the bulk of the old Mcms remained in the cytoplasm, and this defect could not be suppressed by constitutive expression of Cdc6 nor the overexpression of the Cdc14 phosphatase, the only known CDK site phosphatase (Stegmeier and Amon, 2004). This is in marked contrast to the behavior of Swi6, a similarly phosphorylated and exported protein (Geymonat et al., 2004), which efficiently reenters the nucleus upon Cdc14 overproduction. In addition, the transport proteins that move Mcm2-7 into the nucleus are functional under these conditions because the Mcm transport module accumulates in the nucleus in G1 when Yox1 is overexpressed. The simple interpretation of these results is that the Mcms need to be newly transcribed each cell cycle to efficiently accumulate in the nucleus. This is also the case for another pre-RC component, Cdc6, which must be newly transcribed and translated each cycle, because it is degraded in late G1 when CDKs are activated (Piatti et al., 1995, 1996; Elsasser et al., 1999; Drury et al., 2000). The importance of the timely transcription of these pre-RC factors is emphasized by the large increase in chromosome instability that is seen when Yox1 is overexpressed.
Because the old Mcms are not competent to enter the nucleus, they must be degraded to produce the dramatic shift in localization that is observed. Our experiments show that when the transcription of MCM2-7 is blocked by overexpression of Yox1, the old Mcm3 is degraded as the cells pass from mitosis to G1. Consistent with this, Mcm3 is ubiquitinated specifically in G2/M and this ubiquitinated form of Mcm3 disappears as cells exit mitosis (Cheng et al., 2002). This correlates with the timing of the degradation of the old Mcm3 in our experiments. We have also shown that the levels of Mcm3 and Mcm4 are elevated in cells lacking the ubiquitin-activating enzyme Uba1 and mutations in an F-box protein, Met30, lead to elevated levels of Mcm4 (Su et al., 2005). These data suggest that ubiquitination and degradation via the Skp1, Cul1, Skp2 (SCF) complex is an important step in eliminating the old Mcm proteins.
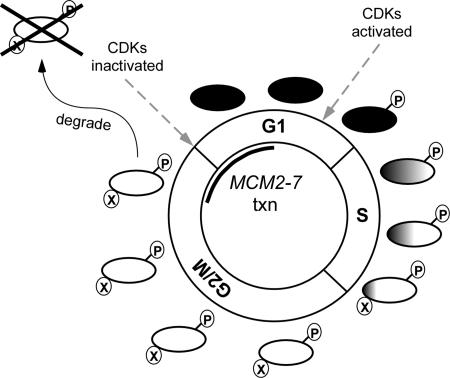
We have shown that neither elimination of CDK activity nor the overproduction of the CDK site phosphatase (Cdc14) is sufficient to permit efficient nuclear reentry of the Mcms. Rather, there is an absolute requirement for nascent transcription and translation for nuclear entry of the Mcms at M/G1. At the same time as the new Mcm proteins are made, the old Mcms are selectively degraded. These results indicate that there are at least two distinct populations of Mcm2-7 complexes; the nascent population present during G1 that has not experienced high CDK activity and is able to accumulate in the nucleus, and the old pool that has been phosphorylated by CDK, exported from the nucleus and targeted for degradation. This leads to an alternative model for the regulation of Mcm2-7 localization that is illustrated in Figure 8. MCM2-7 are transcribed at M/G1 as represented by the black bar inside the cell cycle diagram. As cells exit mitosis, these newly synthesized Mcms accumulate in the nucleus (black ovals) and assemble into pre-RCs during a period of low CDK activity. CDKs are activated toward the end of G1 leading to the phosphorylation of the Mcms (P), which triggers their export from the nucleus (white ovals) progressively throughout S phase as they disassociate from the chromatin. These old Mcms are prevented from reaccumulating in the nucleus due in part to the high CDK activity in mitosis as well as a further unknown modification (X) that cannot be reversed by excess Cdc14 phosphatase. This population is also targeted for degradation through a ubiquitin-mediated pathway. The degradation of the old Mcms and the timely and coordinated transcription and translation of MCM2-7 when CDK activity is low ensures that there is a nascent pool of largely unmodified pre-RC components that can associate with each other and be efficiently transported into the nucleus to form the pre-RC. The simple logic of making a fresh pool of active, unmodified protein just before it is needed, rather than restoring function to inactive proteins dispersed throughout the cell leads us to speculate that this strategy may be a common one.
Figure 8.
Regulation of Mcm2-7 protein localization. See text for detailed discussion of model. The ovals are the Mcm2-7 complexes at various times during the cell cycle. Black filled ovals indicate Mcms are predominately nuclear, white filled ovals indicate Mcms are predominately cytoplasmic, and the ovals with a gradient indicate that only a fraction of the total Mcm population is nuclear. The P indicates phosphorylation, and the X indicates an unknown modification of the Mcms. The timing of MCM2-7 transcription is shown inside the cell cycle diagram. Dashed arrows point to the times when CDKs are activated and inactivated.
The components of the pre-RC are highly conserved, and the assembly of the pre-RC is similarly regulated by CDK activity in higher eukaryotes. In human cells, the CDC6 and MCM2-7 genes are also coordinately transcribed, with most peaking in G1/S (Tsuruga et al., 1997a,b; Hateboer et al., 1998; Leone et al., 1998). However, human MCM4 has recently been shown to be transcribed in M phase (Jensen et al., 2006). This is particularly intriguing because Mcm4 is the subunit whose CDK-dependent phosphorylation controls the chromatin association of the entire Mcm2-7 complex (Ekholm-Reed et al., 2004). It would be of interest to know whether nascent transcription of human Mcm4 also plays a role in the control of pre-RC assembly. The same study (Jensen et al., 2006) provided evidence that, across evolution, periodic transcription, CDK-dependent phosphorylation and proteolysis have coevolved to target the same subunits of complexes that carry out cell cycle-specific processes. Our observations offer an example of how these three levels of regulation act together to control formation of the prereplication complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Miles for technical support and the members of our laboratory for helpful discussions. We are also very grateful for use of the Biggins laboratory's fluorescent microscope. We thank J. Diffley, F. Spencer, S. Gasser, J. Li, S. Sedgwick, and B. Stillman for strains, plasmids, and antibodies. This work was supported by National Institutes of Health grant GM-41073 (to L.B.).
Abbreviations used:
- CDK
cyclin-dependent kinase
- ECB
early cell cycle box
- Mcm
minichromosome maintenance protein
- ORC
origin recognition complex
- pre-RC
prereplication complex.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0792) on February 21, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Aparicio O. M., Weinstein D. M., Bell S. P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Broek D., Bartlett R., Crawford K., Nurse P. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature. 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- Cheng I. H., Roberts L. A., Tye B. K. Mcm3 is polyubiquitinated during mitosis before establishment of the pre-replication complex. J. Biol. Chem. 2002;277:41706–41714. doi: 10.1074/jbc.M205793200. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Diffley J. F., Nasmyth K. A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Dalton S., Whitbread L. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc. Natl. Acad. Sci. USA. 1995;92:2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L. S., Perkins G., Diffley J. F. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- Ekholm-Reed S., Mendez J., Tedesco D., Zetterberg A., Stillman B., Reed S. I. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S., Chi Y., Yang P., Campbell J. L. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch M. J., Donato J. J., Tye B. K. Mcm7, a subunit of the presumptive MCM helicase, modulates its own expression in conjunction with Mcm1. J. Biol. Chem. 2003;278:25408–25416. doi: 10.1074/jbc.M300699200. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Wells G. P., Smerdon S. J., Sedgwick S. G. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol. Cell Biol. 2004;24:2277–2285. doi: 10.1128/MCB.24.6.2277-2285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. M., Morreale R. J., Ozaydin B., Derisi J. L., Li J. J. Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol. Biol. Cell. 2006;17:2401–2414. doi: 10.1091/mbc.E05-11-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G., Wobst A., Petersen B. O., Le Cam L., Vigo E., Sardet C., Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy K. M., Clark C. D., Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990;4:2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- Jensen L. J., Jensen T. S., de Lichtenberg U., Brunak S., Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- Kaplan D. L., Davey M. J., O'Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- Labib K., Diffley J.F.X., Kearsey S. E. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- Labib K., Kearsey S. E., Diffley J. F. Mcm2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol. Biol. Cell. 2001;12:3658–3667. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., Tercero J. A., Diffley J. F. Uninterrupted Mcm2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Lee J. K., Hurwitz J. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki Y., Tye B. K. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol. Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G., DeGregori J., Yan Z., Jakoi L., Ishida S., Williams R. S., Nevins J. R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D. T., Hodson J. A., Forsburg S. L. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J. Cell Sci. 1999;112:559–567. doi: 10.1242/jcs.112.4.559. [DOI] [PubMed] [Google Scholar]
- Liku M. E., Nguyen V. Q., Rosales A. W., Irie K., Li J. J. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2-7 complex prevents chromosomal rereplication. Mol. Biol. Cell. 2005;16:5026–5039. doi: 10.1091/mbc.E05-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. P., Jentsch S., Varshavsky A. UBA 1, an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny C. J., Partridge J. F., Mikesell G. E., Creemer D. P., Breeden L. L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- Nguyen V. Q., Co C., Irie K., Li J. J. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- Nguyen V. Q., Co C., Li J. J. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Noton E., Diffley J. F. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol. Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Piatti S., Bohm T., Cocker J. H., Diffley J.F.X., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Piatti S., Lengauer C., Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T., Miles S., GuhaThakurta D., Jemilo D., Breeden L. L. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002;16:3034–3045. doi: 10.1101/gad.1034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Pasero P., Gasser S. M. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J., Breeden L. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Su N. Y., Flick K., Kaiser P. The F-box protein Met30 is required for multiple steps in the budding yeast cell cycle. Mol. Cell Biol. 2005;25:3875–3885. doi: 10.1128/MCB.25.10.3875-3885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Diffley J. F. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp D., Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tanny R. E., MacAlpine D. M., Blitzblau H. G., Bell S. P. Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol. Biol. Cell. 2006;17:2415–2423. doi: 10.1091/mbc.E05-11-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruga H., Yabuta N., Hosoya S., Tamura K., Endo Y., Nojima H. HsMCM 6, a new member of the human MCM/P1 family encodes a protein homologous to fission yeast Mis5. Genes Cells. 1997a;2:381–399. doi: 10.1046/j.1365-2443.1997.1290327.x. [DOI] [PubMed] [Google Scholar]
- Tsuruga H., Yabuta N., Hashizume K., Ikeda M., Endo Y., Nojima H. Expression, nuclear localization and interactions of human MCM/P1 proteins. Biochem. Biophys. Res. Commun. 1997b;236:118–125. doi: 10.1006/bbrc.1997.6865. [DOI] [PubMed] [Google Scholar]
- Wilmes G. M., Archambault V., Austin R. J., Jacobson M. D., Bell S. P., Cross F. R. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Komamura Y., Ishimi Y. Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol. Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. R., Tye B. K. Mcm2 and Mcm3 are constitutive nuclear proteins that exhibit distinct isoforms and bind chromatin during specific cell cycle stages of Saccharomyces cerevisiae. Mol. Biol. Cell. 1997;8:1587–1601. doi: 10.1091/mbc.8.8.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.