Abstract
Dual-specificity tyrosine-phosphorylated and regulated kinase (DYRK) proteins are an evolutionarily conserved family of protein kinases, with members identified from yeast to humans, that participate in a variety of cellular processes. DYRKs are serine/threonine protein kinases that are activated by autophosphorylation on a tyrosine residue in the activation loop. The family member DYRK1A has been shown to phosphorylate several cytosolic proteins and a number of splicing and transcription factors, including members of the nuclear factor of activated T cells family. In the present study, we show that DYRK1A autophosphorylates, via an intramolecular mechanism, on Ser-520, in the PEST domain of the protein. We also show that phosphorylation of this residue, which we show is subjected to dynamic changes in vivo, mediates the interaction of DYRK1A with 14-3-3β. A second 14-3-3 binding site is present within the N-terminal of the protein. In the context of the DYRK1A molecule, neither site can act independently of the other. Bacterially produced DYRK1A and the mutant DYRK1A/S520A have similar kinase activities, suggesting that Ser-520 phosphorylation does not affect the intrinsic kinase activity on its own. Instead, we demonstrate that this phosphorylation allows the binding of 14-3-3β, which in turn stimulates the catalytic activity of DYRK1A. These findings provide evidence for a novel mechanism for the regulation of DYRK1A kinase activity.
INTRODUCTION
DYRK1A belongs to a family of conserved protein kinases called dual-specificity tyrosine-phosphorylated and regulated kinase (DYRK), within the CMGC group (CDK, MAPK, GSK, and CLK families) of the eukaryote kinome. DYRK family members share a conserved kinase domain and an adjacent DYRK-homology domain, or DH-box (DDDNXDY), but they differ in their N- and C-terminal regions. From a phylogenetic viewpoint, the family can be classified into two subfamilies: a group of cytosolic DYRK proteins, which includes Schizosaccharomyces pombe Pom1p, Caenorhabditis elegans mbk-2, Drosophila melanogaster dDYRK2 and DYRK3, and vertebrate DYRK2, DYRK3, and DYRK4; and a subfamily of DYRKs that are considered mostly nuclear proteins, and which includes Saccharomyces cerevisiae Yak1p, Dictyostelium discoideum YakA, C. elegans mbk-1, D. melanogaster minibrain, and vertebrate DYRK1A and DYRK1B.
The mammalian DYRK1A is ubiquitously expressed in adult and fetal tissues (Guimera et al., 1999; Okui et al., 1999) and shows a specific expression pattern in the central nervous system and during neurogenesis (Hammerle et al., 2002; Marti et al., 2003; Wegiel et al., 2004). Transgenic mice carrying extra copies of the gene exhibit learning defects and motor abnormalities (Smith et al., 1997; Altafaj et al., 2001), and it has been recently described that overexpression of Dyrk1A during mouse embryonic development leads to vascular defects (Arron et al., 2006). Heterozygous Dyrk1A mice also present a noticeable phenotype, with region-specific brain alterations (Fotaki et al., 2002). All these data strongly support the idea that Dyrk1A is a dosage-sensitive gene. This, together with its mapping to chromosome 21q22.2, its overexpression in Down syndrome fetal brains (Guimera et al., 1999), and the phenotype of the DYRK1A transgenic mouse models, has drawn special attention on this gene as a likely contributor to some of the pathological traits observed in chromosome 21 trisomy.
DYRK1A harbors, in addition to the conserved catalytic kinase domain, two nuclear localization signals (NLSs): a classical bipartite NLS at the N terminus and a complex NLS within the catalytic domain (Alvarez et al., 2003). The kinase domain is followed by a PEST domain and then by a repeat of histidines that targets DYRK1A to the splicing factor compartment (Alvarez et al., 2003). DYRK1A has been defined as a dual-specificity kinase because of its ability to autophosphorylate on Tyr and Ser/Thr residues. Moreover, in exogenous substrates it can only phosphorylate Ser and Thr residues that occur within a consensus phosphorylation sequence, RPX(S/T)P, and with a preference for proline in the +1 position (Himpel et al., 2000, 2001). Substrates of DYRK1A, some of them awaiting confirmation in vivo, have been identified among different classes of proteins. These include transcription factors such as Forkhead in rhabdomyosarcoma (FKHR) (Woods et al., 2001), cAMP response element-binding protein (Yang et al., 2001), Gli1 (Mao et al., 2002), and nuclear factor of activated T cells (NFAT) family members (Arron et al., 2006; Gwack et al., 2006); several splicing factors, including cyclin L2 (de Graaf et al., 2004) and SF3b/SAP155 (de Graaf et al., 2006); and some cytosolic molecules such as dynamin (Chen-Hwang et al., 2002) and the enzyme glycogen synthase (Skurat and Dietrich, 2004). DYRK1A has also been reported to potentiate the neural growth factor-induced differentiation of PC12 cells through its interaction with components of the signaling cascade Ras-BRaf-MEK1, but this effect is independent of its kinase activity (Kelly and Rahmani, 2005).
All members of the DYRK family are characterized by a conserved Tyr-X-Tyr motif in the activation loop of the catalytic domain (Becker and Joost, 1999). Phosphorylation of the second Tyr residue of this motif (Tyr-321 in DYRK1A) is essential for the kinase activity of all the DYRK members reported so far (Himpel et al., 2001; Li et al., 2002; Lochhead et al., 2003). The mechanism of activation of DYRKs resembles that of the mitogen-activated protein kinase (MAPK) family, although, unlike MAPKs, Tyr phosphorylation in the DYRK activation loop is an autophosphorylation event and does not involve an upstream activating kinase (Himpel et al., 2001). A recent report on the Drosophila DYRKs minibrain and dDYRK2 shows that autophosphorylation in the activation loop of these kinases is an intramolecular event, mediated by a transitional intermediate form during translation (Lochhead et al., 2005). The Tyr-kinase activity is lost once the protein is fully translated, and the mature kinase can only phosphorylate Ser/Thr residues. Assuming this mechanism to be general for all DYRKs, the autophosphorylation of the activation loop in DYRK1A would be constitutive. However, some DYRK1A functions are clearly dose and activity dependent (Arron et al., 2006; Gwack et al., 2006), and it is therefore likely that the activity of DYRK1A will be regulated by additional mechanisms.
There are several ways in which protein kinases can stabilize an active conformation, in addition to the most commonly used mechanism of phosphorylation within the activation loop. These include phosphorylation/dephosphorylation events in parts of the catalytic domain other than the activation segment or in other domains, the use of separate regulatory subunits, and binding to other regulatory proteins. One such group of binding proteins that are known to modulate the activity of some of their kinase targets is the 14-3-3 protein family. The 14-3-3 family is a highly conserved and widely expressed group of 28- to 31-kDa molecules that naturally assemble as homo- or heterodimers (Jones et al., 1995). In general, 14-3-3 binding depends on phosphorylated Ser or Thr residues within the target protein. Studies with phosphopeptide libraries have identified two preferred binding motifs for 14-3-3: RSXpSXP (mode I) and RXXXpSXP (mode II), where pS is a phospho-Ser residue (Yaffe et al., 1997). More recently a mode III consensus binding sequence has also been proposed: the C-terminal binding motif pSX1-2-COOH (Ganguly et al., 2005). However, a number of proteins associate with 14-3-3 through other phosphorylated sequences or even through nonphosphorylated motifs (for a recent review, see Aitken, 2006). Binding by 14-3-3 proteins has four major biological effects on target proteins: 1) inhibition or activation of catalytic activities, 2) regulation of protein stability, 3) modulation of subcellular localization, and 4) contribution to the formation of macromolecular complexes. The variety of 14-3-3 partners accounts for the ability of this family of proteins to modulate many signaling pathways and cellular processes (Dougherty and Morrison, 2004).
We identified 14-3-3β during a screen for DYRK1A-interacting proteins that might modulate its activity, and decided to explore this interaction further. In the present study we show that phosphorylation of Ser-520 in DYRK1A is required for 14-3-3β binding. DYRK1A autophosphorylates Ser-520 through an intramolecular reaction. Finally, we demonstrate that 14-3-3β binding significantly increases the catalytic activity of DYRK1A.
MATERIALS AND METHODS
Plasmids
The expression plasmids pHA-DYRK1A and pGFP-DYRK1A have been described previously (Alvarez et al., 2003; Marti et al., 2003); these plasmids, respectively, encode N-terminal hemagglutinin epitope (HA)-tagged and enhanced green fluorescent protein (EGFP)-tagged fusion protein versions of the 754-amino acid alternative spliced isoform of human DYRK1A. To create the plasmid expressing the 763-amino acid HA-tagged DYRK1A isoform (from hereon identified as +9), we cloned a BglII–SacI fragment (amino acids 1–208) from the plasmid pEGFP-DYRK1A (+9) and a SacI–XbaI fragment (amino acids 209–763) from the plasmid pHA-DYRK1A into the BamHI and XbaI sites of pCDNA-HA. The plasmid pHA-DYRK1A/S520A and the kinase-inactive mutants pHA-DYRK1A/K179R and pHA-DYRK1A/Y310,312F were generated by site-directed mutagenesis and verified by DNA sequencing. Plasmids expressing glutathione S-transferase (GST)-fusion proteins for wild-type DYRK1A (wt) and DYRK1A/K179R were generated by subcloning the fragments BamHI-SacI (amino acids 1–199) and SacI-NotI (amino acids 200–754) from their corresponding pHA-DYRK1A plasmids into the BamHI and NotI sites of pGEX-4T3 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). To generate the GST-DYRK1A/S520A–expressing plasmid, a SacI–NotI fragment (amino acids 200–754) in pGST-DYRK1A was replaced by the equivalent fragment from pHA-DYRK1A/S520A, which contains the point mutation. Plasmid pEGFP-DYRK1A/1–167 was made by subcloning a BamHI–SalI PCR fragment (amino acids 1–167) into pEGFP BglII-XhoI. Plasmids pEGFP-DYRK1A (+9) and pGST-DYRK1A/ΔN (amino acids 149–763) (Himpel et al., 2001) were kindly provided by W. Becker (Medizinische Fakultaet der RWTH, Aachen, Germany).
The human 14-3-3β open reading frame was amplified by polymerase chain reaction (PCR) from the pACTII-14-3-3β plasmid isolated from the cDNA library used in the yeast two-hybrid screen (human fetal brain library in pACTII; Clontech, Mountain View, CA). The forward primer, flanked with a BamHI restriction site, was 5′-ggatcccaatggataaaaagtgag-3′, and the reverse primer 5′-tgcacgatgcacagttgaagt-3′. The PCR product was cloned into pGEM-Teasy (Promega, Madison, WI), sequenced, and then inserted in-frame into the BamHI and EcoRI sites of pGEX-2T (GE Healthcare).
Antibodies
The anti-DYRK1A antibody used in this study was raised by immunizing rabbits with a purified GST-fusion protein making up the last 144 amino acids of DYRK1A. Serum specificity was tested by competition experiments and by comparing the antibody reactivity against extracts from Dyrk1Awt and Dyrk1A−/− mouse embryos, kindly provided by M. L. Arbones (CRG, Barcelona, Spain). The antibody was affinity purified using the GST orientation kit (Pierce Chemical, Rockford, IL), according to the manufacturer's instructions. To generate the specific anti-DYRK1A phospho-S520 antibody, rabbits were immunized with the phosphopeptide SNSGRARpSDPTHQHR (where pS is a phospho-Ser residue) conjugated to keyhole limpet hemocyanine. The serum obtained was purified on an affinity column by using the SulfoLink kit (Pierce Chemical), following the manufacturer's recommendations.
Commercially available antibodies used in this study included a monoclonal antibody (mAb) (HA.11) to the HA-epitope tag (Covance, Princeton, NJ), an anti-phosphotyrosine antibody (PY20; BD Biosciences PharMingen, San Diego, CA), a mAb against green fluorescent protein (GFP) proteins (JL-8; BD Biosciences Clontech), a pan-14-3-3 antibody (H-8; Santa Cruz Biotechnology, Santa Cruz, CA), a phospho-p44/42 MAPK (Thr202/Tyr204) mAb (Cell Signaling Technology, Beverly, MA), and a β-tubulin antibody (Chemicon International, Temecula, CA). Horseradish peroxidase (HRP)-conjugated mAb against GST was from Santa Cruz Biotechnology. HRP-conjugated rabbit anti-mouse and goat anti-rabbit antibodies (Dako Denmark A/S, Glostrup, Denmark) were used as secondary antibodies for immunoblotting. Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse was from Southern Biotechnology Associates (Birmingham, AL).
Cell Culture and Transfection
U2-OS cells were purchased from the European Collection of Cell Cultures (Wiltshire, United Kingdom), and human embryonic kidney (HEK)293 cells were kindly provided by M. Palacín (University of Barcelona, Barcelona, Spain). Both cell lines were maintained at 37°C in DMEM supplemented with 10% fetal calf serum (FCS) and antibiotics. Transient transfections were performed using the calcium phosphate method, and cells were processed 48 h after transfection. Treatment of U2-OS cells with SB216763 (Sigma-Aldrich, St. Louis, MO), or dimethyl sulfoxide (DMSO) as vehicle, was carried out for 8 h at the concentrations indicated.
The PC12 cell line was a gift from J. Comella (University of Lleida, Lleida, Spain) and was maintained in DMEM supplemented with 6% FCS, 6% horse serum, and antibiotics. For fibroblast growth factor (FGF) stimulation, PC12 cells were serum starved (DMEM supplemented with antibiotics) during 20 h and then treated with 100 ng/ml human bFGF (PeproTech EC, London, United Kingdom) for the times indicated.
Immunofluorescence
U2-OS cells growing on coverslips were transfected with different DYRK1A-expressing constructs. Forty-eight hours after transfection, cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and processed for immunofluorescence as described previously (Alvarez et al., 2003). Images were captured with a Leica DFC 300FX camera mounted on a Leica DMR microscope (Leica, Wetzlar, Germany).
Cell Extracts and Immunoblotting
Whole-cell extracts were prepared in SDS-buffer (25 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 1% SDS). For GST pull-down and immunoprecipitation assays, soluble extracts were prepared by resuspending cell pellets for 20 min at 4°C in lysis buffer A (50 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, protease inhibitor cocktail [Roche Diagnostics, Mannheim, Germany], and phosphatase inhibitors [2 mM sodium orthovanadate, 30 mM sodium pyrophosphate, and 25 mM sodium fluoride]), followed by centrifuging for 10 min at 13,000 × g at 4°C.
For immunoblotting analysis samples were resolved by SDS-PAGE, transferred onto a nitrocellulose membrane (Hybond C; GE Healthcare), and blocked with 10% skimmed milk in Tris-buffered saline (TBS) (10 mM Tris-HCl, pH 7.5, and 100 mM NaCl) containing 0.1% Tween 20 (TBS-T). Membranes were incubated with primary antibodies (in 5% skimmed milk in TBS-T) overnight at 4°C, except for PY20 antibody, when bovine serum albumin (BSA) replaced skimmed milk for both blocking and antibody incubation. After washing with TBS-T, membranes were incubated for 45 min at room temperature (RT) with the appropriate secondary antibodies (in 5% skimmed milk in TBS-T) and then washed again with TBS-T. Detection was by enhanced chemiluminescence with Supersignal West Pico (Pierce Chemical). For detection of pS520 in the endogenous DYRK1A protein, ECL Plus (GE Healthcare) was used to increase sensitivity. Chemiluminescence was determined with a LAS-3000 image analyzer (Fuji PhotoFilm, Tokyo, Japan). Quantification of data was performed using Image Gauge software version 4 (Fuji PhotoFilm).
GST-Fusion Protein Expression in Bacteria
GST-fusion expressing constructs were transformed into Escherichia coli BL21(DE3)pLysS (Stratagene, La Jolla, CA). Protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactoside for 3 h at 37°C for GST-14-3-3β and for 8 h at 20°C for GST-DYRK1A. Cells were lysed in lysis buffer B (10 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, and a protease inhibitor cocktail). Bacterial lysates were incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 45 min at RT and washed four times with lysis buffer B. GST-DYRK1A fusion proteins were eluted with 10 mM reduced glutathione (Sigma-Aldrich) in 50 mM Tris-HCl, pH 8, and dialyzed against a buffer containing 50 mM HEPES, pH 7.4, 150 mM NaCl, and 2 mM EDTA.
Pull-Down Assays
Soluble cell lysates were incubated overnight at 4°C with 10 μg of unfused GST or GST-14-3-3β immobilized on glutathione-Sepharose beads that had been previously equilibrated in lysis buffer A. After binding, beads were washed four times with lysis buffer A plus 30 mM sodium pyrophosphate, and the bound protein was eluted by boiling samples for 5 min in SDS-buffer. Samples were resolved by 8 or 10% SDS-PAGE and proteins were detected by immunoblotting.
Phosphatase Treatment
Cells were lysed in phosphatase buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 2 mM MgCl2, 1% NP-40, 2 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail) in the absence of phosphatase inhibitors. Alkaline phosphatase (Sigma-Aldrich) was added to lysates at a final concentration of 400 U/ml (0.2 U/μg protein), and the reaction mixes were incubated for 30 min at 30°C. To stop phosphatase activity, sodium pyrophosphate was added to the lysates at a final concentration of 25 mM, and samples were processed.
Immunoprecipitation
Soluble cell extracts were incubated overnight at 4°C with protein G-Sepharose beads (GE Healthcare) prebound with 5 μg of anti-HA antibody. Beads were washed four times with lysis buffer A, adding 0.1% NP-40 for the two initial washes. For immunoprecipitation of endogenous DYRK1A protein from PC12 cells, a soluble cell extract containing 2.5 mg of total protein was prepared as described above. The lysate was first incubated overnight at 4°C with 10 μg of the anti-DYRK1A antibody, and it was then incubated with protein A-Sepharose beads (GE Healthcare) for 2 h at 4°C. The immunoprecipitates were then washed with lysis buffer A containing 30 mM sodium pyrophosphate. Samples were resolved by SDS-PAGE and analyzed by immunoblotting and/or used for in vitro kinase assay as described below.
Kinase Assays
Kinase activity of DYRK1A proteins was determined with the peptide substrate DYRKtide (Himpel et al., 2000). To determine the catalytic activity of GST-DYRK1A proteins, 50 ng of the purified recombinant protein was incubated for 10 min at 20°C in 50 μl of phosphorylation buffer (50 mM HEPES, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, and 1 mM dithiothreitol) containing 200 μM DYRKtide and 100 μM [γ-32P]ATP (0.2 × 10−3 μCi/pmol). Reaction aliquots were dotted onto P81 Whatman paper, and, after washing extensively with 5% orthophosphoric acid, counts were determined in a liquid scintillation counter. For assays of HA-DYRK1A proteins, anti-HA immunocomplexes were incubated for 20 min at 30°C in 20 μl of phosphorylation buffer, in the presence of 200 μM DYRKtide and 50 μM [γ-32P]ATP (1 × 10−3 μCi/pmol), and 32P incorporation was determined as described above. Each data point was determined in triplicate and normalized against the amount of HA–DYRK1A protein present in the immunocomplexes, as quantified by immunoblot with anti-HA. Quantifications were done using the Image Gauge software. The data were evaluated for statistical significance with the two-tailed Student's test for independent samples, and differences were considered significant at p ≤ 0.05.
RESULTS
Phosphorylation of DYRK1A on Serine 520 Is Required for 14-3-3β Binding
To identify DYRK1A interacting proteins, we performed a yeast two-hybrid screen of a human fetal brain cDNA library, by using the human DYRK1A full-length open reading frame (754-amino acid isoform) as the bait. One of the positive clones encoded the protein 14-3-3β, one of the seven isoforms of the 14-3-3 family of proteins in mammals.
We confirmed DYRK1A and 14-3-3 interaction in mammalian cells by GST pull-down assays. HEK293 cells were transfected with a HA-tagged DYRK1A expression plasmid, and soluble lysates were incubated with recombinant GST-14-3-3β. Bound proteins were analyzed by immunoblotting with an anti-HA antibody. As shown in Figure 1, DYRK1A was pulled down by the GST-14-3-3β fusion protein but not by unfused GST. Given that most reported interactions of 14-3-3 are mediated by phosphorylated Ser or Thr residues present in their target proteins, we examined whether phosphorylation of DYRK1A was required for binding. We treated soluble extracts prepared from cells overexpressing HA-DYRK1A with alkaline phosphatase, and we pulled down with GST-14-3-3β. 14-3-3 binding was almost abolished after phosphatase treatment (Figure 1A, lane 2), but it was maintained when the extract was pretreated with the phosphatase inhibitor sodium pyrophosphate (Figure 1A, lane 3). This result indicates that 14-3-3β interaction is dependent on the phosphorylation status of DYRK1A.
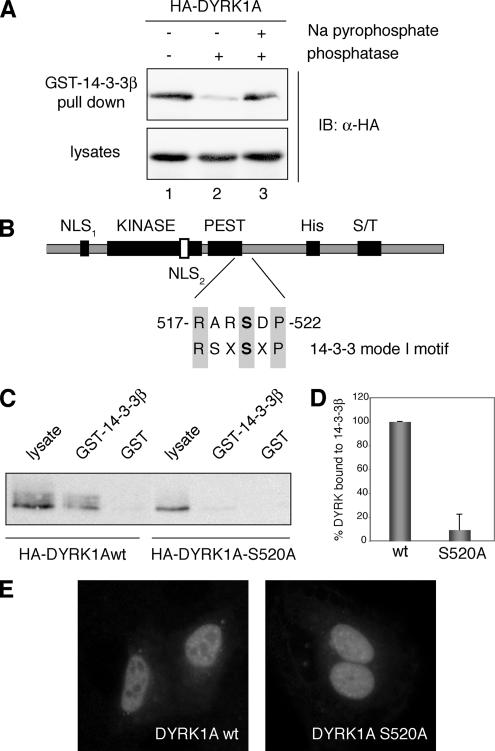
Figure 1.
Phosphorylation of Ser-520 on DYRK1A mediates its interaction with the protein 14-3-3β. (A) Lysates from HEK293 cells transiently transfected with pHA-DYRK1A were incubated for 30 min at 30°C in phosphatase buffer (lane 1) supplemented with alkaline phosphatase (lane 2) or with alkaline phosphatase and sodium pyrophosphate (lane 3) as indicated. After incubation with GST-14-3-3β immobilized on glutathione-Sepharose beads, bound protein was detected by immunoblotting (IB) with anti-HA antibody (top). An immunoblot of cell lysates representing 10% of the inputs is shown (bottom). (B) Schematic diagram of DYRK1A showing the domain structure: KINASE, kinase catalytic domain; PEST, PEST domain; His, histidine-repeat; and S/T, Ser/Thr-rich region. The sequence and the position of the putative 14-3-3 binding motif in DYRK1A are indicated and compared with a mode I 14-3-3 binding consensus sequence. The putative phosphorylated Ser residue is marked in bold. (C) HEK293 cells were transfected with pHA-DYRK1Awt and pHA-DYRK1A/S520A, and soluble lysates were incubated with unfused GST or GST-14-3-3β. Samples were analyzed by SDS-PAGE (10%) followed by immunoblotting with anti-HA antibody. Lysate represents 10% of the input. (D) The histogram presents data from three independent experiments in which the ratio bound/input in DYRK1Awt was arbitrarily set as 100. Data are expressed as a percentage of the mean value. Error bars represent SEM (standard error of the mean). (E) Subcellular localization of HA-DYRK1Awt and the mutant HA-DYRK1A/S520A, expressed in U2-OS cells, by indirect immunofluorescence analysis with anti-HA antibody followed by a FITC-conjugated anti-mouse secondary antibody.
We next searched DYRK1A for the presence of the known consensus motifs for 14-3-3 binding: RSXpSXP (mode I) or RXXXpSXP (mode II), where pS represents phosphoserine (Yaffe et al., 1997). An initial yeast two-hybrid analysis of the 14-3-3β interaction with a panel of DYRK1A deletion mutants pointed to the involvement of the DYRK1A C-terminal region (Supplemental Figure S1). In the mapped fragment, a putative 14-3-3 mode I binding motif, 517-RARSDP-522, was identified close to the PEST domain of DYRK1A (Figure 1B). This motif is not a perfect match because of the presence of Ala instead of Ser at position −2 relative to the phospho-Ser residue; however, the Arg at position −1 satisfies the preference for an aromatic or positively charged amino acid at this position (Yaffe et al., 1997). To determine whether the predicted motif was required for 14-3-3β binding, we generated an HA-tagged version of DYRK1A in which the potentially phosphorylated residue Ser-520 is replaced by Ala (HA-DYRK1A/S520A). The mutant form of DYRK1A did not show any detectable interaction with GST-14-3-3β in pull-down assays (Figure 1C, representative pull-down; Figure 1D, quantification of the data from three independent experiments), which indicates that DYRK1A very likely needs to be phosphorylated on Ser-520 to bind to 14-3-3β and confirms that this residue is located in a critical phospho-Ser recognition motif for 14-3-3.
Binding by 14-3-3 modulates the subcellular localization of many of its targets (Dougherty and Morrison, 2004). To test whether this takes place in DYRK1A, we compared the subcellular localization of exogenously expressed DYRK1Awt and the mutant DYRK1A/S520A in U2-OS cells by indirect immunofluorescence. The staining pattern of DYRK1A/S520A was similar to that of DYRK1Awt, with a clear accumulation in the nucleus and with concentration in nuclear speckles, in addition to a slight cytoplasmic staining (Figure 1E). This is the same pattern we have described previously for DYRK1A (Alvarez et al., 2003), and it suggests that 14-3-3 binding does not induce major changes in the subcellular localization of DYRK1A. However, we cannot rule out that 14-3-3 binding could modulate the localization of DYRK1A under specific conditions, or affect the dynamics of its nucleocytoplasmic transport. Indeed, in Yak1p and Bmh1/2p, the DYRK1A and 14-3-3 homologues in S. cerevisiae, Yak1p nuclear export is induced by glucose through the binding of Bmh1/2p (Moriya et al., 2001).
DYRK1A Is Phosphorylated on Ser-520 In Vivo
To enable us to examine the phosphorylation status of the Ser-520 residue in vivo, we generated an antibody against the DYRK1A sequence spanning Ser-520 and in which this residue was phosphorylated (pS520). To validate the specificity of this antibody, we first performed peptide competition assays. Whereas the immunizing peptide in its phosphorylated form fully competed away the anti-pSer520 signal on immunoblots, no inhibitory effect was observed with the nonphosphorylated version of the same peptide (Figure 2A). In addition, phosphatase treatment eliminated the specific signal detected by the phospho-specific pS520 antibody (Figure 2B), suggesting that the reactivity of this antibody depends on the phosphorylation status of the protein. Finally, we evaluated the specificity of the antibody by examining the reactivity of extracts overexpressing HA-DYRK1Awt and HA-DYRK1A/S520A by immunoblot. Whereas anti-HA antibody detected similar expression levels of both HA-tagged proteins (Figure 2C, lanes 1 and 2), the phospho-specific anti-DYRK1A pS520 antibody recognized DYRK1Awt (Figure 2C, lane 3), but it did not show any reactivity toward DYRK1A/S520A (Figure 2C, lane 4). Moreover, the antibody preferentially recognized bands of lower electrophoretic mobility (Figure 2C, lane 3) that correspond to phosphorylated DYRK1A species, as determined by their sensitivity to phosphatase treatment (Figure 2B). These results demonstrate that the antibody is specific for DYRK1A phosphorylated on Ser-520 and represents a valuable tool for tracking DYRK1A phosphorylation under physiological conditions.
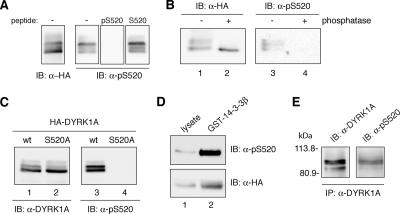
Figure 2.
DYRK1A is phosphorylated on Ser-520 in vivo. (A) Whole-cell extracts from U2-OS cells expressing HA-DYRK1Awt were resolved by 8% SDS-PAGE and analyzed by immunoblot with anti-HA antibody (left) and with anti-pS520 antibody (right). Where indicated, the phosphorylated immunizing peptide (pS520: SNSGRARpSDPTHQHR) or an equivalent unphosphorylated peptide (S520: SNSGRARSDPTHQHR) was added at 25 μg/ml during the incubation period with 1 μg/ml anti-pS520 antibody. (B) Lysates from U2-OS cells transiently transfected with pHA-DYRK1A were incubated for 30 min at 30°C in phosphatase buffer alone (lanes 1 and 3) or supplemented with alkaline phosphatase (lanes 2 and 4), as indicated. Samples were analyzed by SDS-PAGE and immunoblot with anti-HA antibody (lanes 1 and 2) and with the anti-DYRK1A pS520 antibody (lanes 3 and 4). (C) U2-OS cells were transiently transfected with pHA-DYRK1Awt (lanes 1 and 3) and pHA-DYRK1A/S520A (lanes 2 and 4). Whole-cell extracts were resolved by 8% SDS-PAGE and immunoblotted with an anti-HA antibody (lanes 1 and 2) and with the anti-DYRK1A pS520 antibody (lanes 3 and 4). (D) Soluble cell extracts from U2-OS cells expressing HA-DYRK1Awt were incubated with GST-14-3-3β. Lysates, representing 10% of the input, (lane 1) and pulled-down proteins (lane 2) were separated by 8% SDS-PAGE and analyzed by immunoblot with anti-HA antibody (bottom) and with the phospho-specific anti-DYRK1A pS520 antibody (top) to detect the presence of Ser-520–phosphorylated DYRK1A bound to 14-3-3β. (E) The endogenous DYRK1A protein was immunoprecipitated from PC12 cells with anti-DYRK1A antibody. The immunoprecipitate was analyzed by immunoblot (8% SDS-PAGE) with both the anti-DYRK1A and the anti-DYRK1A pS520 antibody. The position of marker proteins (in kilodaltons) is indicated.
The phospho-specific antibody was used to verify that the fraction of DYRK1A that was pulled down by GST-14-3-3β was effectively phosphorylated on Ser-520. As shown in Figure 2D, a marked enrichment in the phosphorylation level of Ser-520 was detected in the 14-3-3β–bound fraction, compared with the proportion of pS520 signal present in the input lysate (Figure 2D, compare lanes 1 and 2). Consistent with this finding, the low mobility DYRK1A bands were preferentially enriched in the bound fraction.
Finally, to rule out the possibility that the detected phosphorylation was an artifact of overexpression, we analyzed the Ser-520 phosphorylation of endogenously expressed DYRK1A. For this, DYRK1A was immunoprecipitated from PC12 cell extracts with an anti-DYRK1A antibody that recognizes the C-terminal region of the protein, and phosphorylation on Ser-520 was then detected by immunoblotting with the anti-DYRK1A pS520 antibody. The phospho-specific antibody clearly detected specific bands of the expected size in the anti-DYRK1A immunoprecipitates (Figure 2E), thus identifying the second in vivo phosphorylation site in DYRK1A, distinct from the previously described Tyr-phosphorylation site in the activation loop (Himpel et al., 2001).
We next considered whether phosphorylation on Ser-520, rather than being constitutive, might be regulated. Because DYRK1A has been shown to participate in signaling pathways induced by neurotrophic factors (Yang et al., 2001; Kelly and Rahmani, 2005), we analyzed DYRK1A Ser-520 phosphorylation in response to growth factor stimulation. PC12 cells were serum deprived and then stimulated with basic fibroblast growth factor (bFGF). Phosphorylation of Ser-520 was monitored by immunoblot analysis with the anti-DYRK1A pS520 antibody and normalized against total DYRK1A expression detected with the anti-DYRK1A antibody (Figure 3A, representative experiment; Figure 3B, quantification of the data from four experiments). The quantification of several independent experiments showed dynamic changes in Ser-520 phosphorylation following the activation wave of the MAPK ERK. After exposure to bFGF for 1 h, the level of S520 phosphorylation tailed off significantly (Figure 3A, lane 6; p < 0.001). We also tested the effect of epidermal growth factor (EGF) stimulation under the same conditions, but we detected no changes in DYRK1A Ser-520 phosphorylation in response to the stimuli (Supplemental Figure S2, B–D). In summary, these results indicate that the levels of phosphorylation in DYRK1A Ser-520 can be modulated in vivo.
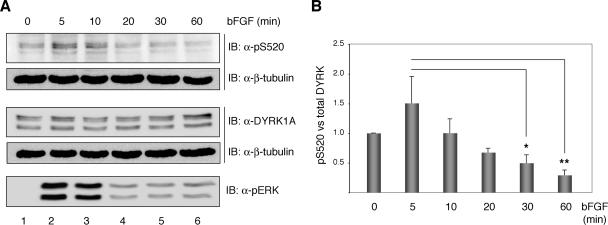
Figure 3.
bFGF modulate the phosphorylation status of DYRK1A Ser-520 in PC12 cells. (A) PC12 cells were serum-deprived for 20 h and then left unstimulated or stimulated with 100 ng/ml bFGF for the times indicated. Total extracts were resolved by 8% SDS-PAGE and analyzed by immunoblot on two independent membranes for the presence of phosphorylated Ser-520 and total DYRK1A. Both membranes were reprobed with anti β-tubulin antibody to confirm equal loading. Phosphorylation of the MAPK extracellular signal-regulated kinase (ERK) was detected by immunoblot with anti-phospho-p44/42 ERK (Thr202/Tyr204) antibody. (B) The chart shows the quantification of the phosphorylation levels of DYRK1A Ser-520 relative to the total amount of DYRK1A over time from the initiation of bFGF stimulation. Unstimulated cells were assigned the baseline value of 1 in each experiment. Data correspond to means ± SEM from four independent experiments. *p ≤ 0.05; **p ≤ 0.001.
DYRK1A Autophosphorylates on Ser-520 through an Intramolecular Mechanism
During the characterization of the anti-DYRK1A pS520 antibody, we surprisingly found that bacterially expressed recombinant GST-DYRK1A was already phosphorylated on Ser-520 (Figure 4A, top, lane 1). Phosphorylation of Ser-520 was detected both in the full-length protein and also in C-terminal–truncated forms that, according to their estimated molecular weights, would be expected to include an intact kinase domain. However, Ser-520 phosphorylation was not detected in the S520A mutant or in a catalytically inactive mutant (DYRK1A/K179R), in which the conserved Lys in the ATP-binding site is replaced by Arg (Figure 4A, top, lanes 2 and 3). In addition, the immunoreactivity with the antibody was abolished when the extracts where treated with phosphatase before the Western blot analysis (Supplemental Figure S3, B). The levels of Tyr-phosphorylation in the DYRK1A/S520A mutant were similar to those in DYRK1Awt (Figure 4A, middle), indicating that the Tyr-autophosphorylation activity of DYRK1A is not affected by the mutation.
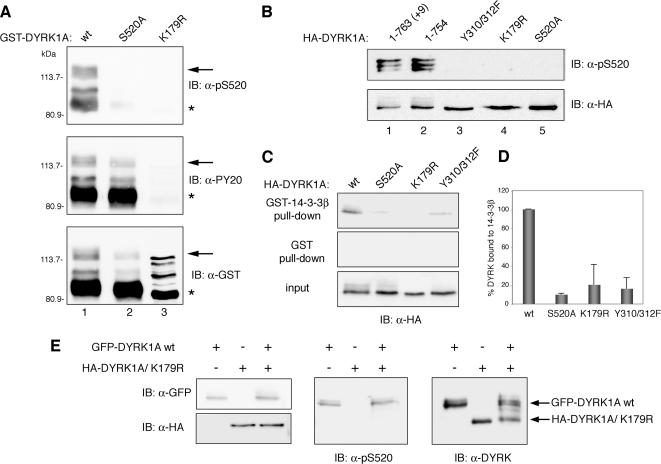
Figure 4.
DYRK1A autophosphorylates Ser-520 through an intramolecular reaction. (A) Bacterially expressed GST fusion proteins of wild-type DYRK1A (wt, lane 1) and the mutants DYRK1A/S520A (lane 2) and DYRK1A/K179R (lane 3) were immunoblotted with anti-DYRK1A pS520 (top), anti-phospho-Tyr (PY20; middle), and anti-GST (bottom) antibodies. The positions of marker proteins (in kilodaltons) are indicated. Arrows indicate full-length GST-fusion proteins, and asterisks indicate truncated GST-fusion proteins lacking the C-terminal (∼150) amino acids). The different patterns in DYRK1Awt, DYRK1AS520A, and DYRK1AK179R detected with the anti-GST antibody are due to differences in phosphorylation levels as shown in Supplemental Figure S3, A. (B) HA-tagged proteins corresponding to the two major isoforms of DYRK1A (lanes 1 and 2) and to distinct catalytically inactive point mutants (lanes 3–5) were overexpressed by transient transfection into U2-OS cells. Phosphorylation of Ser-520 was detected by immunoblotting with the phospho-specific anti-DYRK1A pS520 antibody (top). To control for equal expression of the fusion proteins, membranes were stripped and reprobed with an anti-HA antibody (bottom). (C) U2-OS cells were transfected with pHA-DYRK1Awt, pHA-DYRK1A/S520A, pHA-DYRK1A/K179R, or pHA-DYRK1A/Y310,312F, and soluble lysate fractions were incubated with unfused GST or GST-14-3-3β. Samples were analyzed by SDS-PAGE followed by immunoblotting with anti-HA antibody. Lysate represents 10% of input. (D) Quantification of the relative binding of different DYRK1A mutants to 14-3-3β compared with the fraction bound to DYRK1Awt is shown in the chart, which represents means ± SEM from three independent experiments. (E) A GFP-DYRK1A wild-type fusion protein (GFP-DYRK1Awt) and an HA-tagged DYRK1A kinase-inactive mutant (HA-DYRK1A/K179R) were individually expressed or coexpressed in U2-OS cells and detected by immunoblotting with anti-GFP and anti-HA antibodies, respectively (left). Phosphorylation of Ser-520 was analyzed by immunoblotting with the anti-DYRK1A pS520 antibody (middle). The expression levels of the two DYRK1A fusion proteins were compared by immunoblotting with anti-DYRK1A antibody (right).
We further examined the phosphorylation state of Ser-520 in two kinase-inactive forms of DYRK1A expressed as HA-tagged fusions in U2-OS cells: the mutant in the ATP-binding site (HA-DYRK1A/K179R) and a double mutant in the two Tyr residues of the activation loop (HA-DYRK1A/Y310,312F). Similarly to the bacterially expressed proteins, we could not detect phosphorylation of Ser-520 in these inactive mutants (Figure 4B, lanes 3 and 4), confirming that phosphorylation of Ser-520 in DYRK1A is an autophosphorylation event. Consistent with this result, the kinase-inactive mutants of DYRK1A were unable to bind 14-3-3β protein (Figure 4C, representative pull-down; Figure 4D, quantification of three independent experiments). We also found no differences in the levels of Ser-520 phosphorylation between the two major DYRK1A alternative spliced isoforms: the 763-amino acid form (HA-DYRK1A +9) and the 754-amino acid isoform (HA-DYRK1A) (Figure 4B, lanes 1 and 2). These results suggest that phosphorylation of DYRK1A Ser-520 is an autophosphorylation event and is not mediated by the activity of an upstream kinase.
To determine whether the autophosphorylation of DYRK1A on Ser-520 is mediated by an inter- or intramolecular mechanism, we took advantage of the difference in size of two tagged DYRK1A proteins. We coexpressed the wild-type form of DYRK1A fused to GFP (GFP-DYRK1Awt) and an HA-tagged inactive mutant (HA-DYRK1A/K179R), predicting that if the phosphorylation was intermolecular, the kinase inactive DYRK1A would be phosphorylated by the active version. As shown in Figure 4E, the inactive mutant HA-DYRK1A/K179R was not phosphorylated on Ser-520 when coexpressed with the active GFP-tagged enzyme, a finding consistent with a cis- or intramolecular mechanism. In summary, these results demonstrate that phosphorylation of DYRK1A on Ser-520 is an intramolecular autophosphorylation event.
Phosphorylation of Ser-520 Modulates DYRK1A Kinase Activity
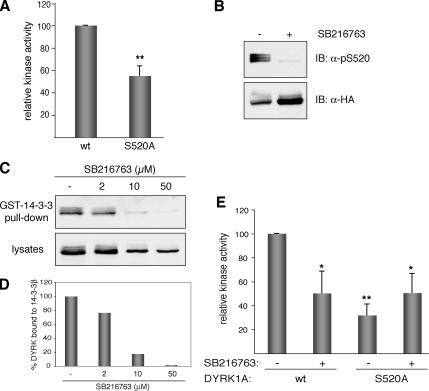
To analyze the possible effect of 14-3-3 binding on the activity of DYRK1A, we determined the kinase activity of the wild-type DYRK1A and of the Ser-520 mutant, which does not interact with 14-3-3β. Activity was measured in in vitro kinase assays with DYRKtide peptide as substrate and anti-HA immunocomplexes obtained from transiently transfected cells as the enzyme source. The S520A mutant showed a significant reduction in the catalytic activity, maintaining only ∼50% of the activity of DYRK1Awt (Figure 5A). To exclude the possibility that this impaired activity resulted from the amino acid substitution rather than the phosphorylation itself, we blocked in vivo the phosphorylation of Ser-520 in DYRK1Awt by using the compound SB216763. This small molecule was first identified as a selective ATP-competitive inhibitor of glycogen synthase kinase (GSK)-3 and was later reported to inhibit DYRK1A activity in vitro at slightly higher concentrations (Murray et al., 2004). Cells expressing HA-DYRK1Awt were treated with SB216763, and the lysates were analyzed by immunoblot. Total DYRK1A protein levels were almost unaffected by the treatment (Figure 5B, IB: HA), but phosphorylation on Ser-520 was markedly reduced in SB216763-treated cells (Figure 5B, IB: pS520). Because GSK-3 does not phosphorylate DYRK1A on Ser-520 (Supplemental Figure S4), the loss of Ser-520 phosphorylation after SB216763 treatment can be ascribed to the inhibition of the kinase activity of DYRK1A. In agreement with the decreased Ser-520 phosphorylation, the levels of HA-DYRK1A pulled down by GST-14-3-3β were reduced in a dose-dependent manner (Figure 5C, representative experiment; 5D, quantification of the data). Supporting these data, the two effects can be reproduced with other DYRK1A kinase inhibitor as shown in Supplemental Figure S5. At this point, and considering also that DYRK1A has been proposed to act as a GSK-3 priming kinase (Cohen and Frame, 2001), it is extremely important to note the risk of attributing phosphorylation events to the kinase GSK-3 based in the use of the inhibitor SB216763, because we have clearly confirmed that this compound also strongly inhibits the activity of DYRK1A in vivo.
Figure 5.
Kinase activity of DYRK1A is modulated by the phosphorylation state of Ser-520. (A) US-OS cells were transfected with pHA-DYRK1Awt and the mutant pHA-DYRK1A/S520A. To determine the kinase activity of DYRK1A, tagged proteins were isolated by immunoprecipitation with anti-HA antibody, and immunocomplexes were subjected to in vitro kinase assay with DYRKtide as substrate. Equal amounts of HA-tagged proteins were used in all cases. The data represent means ± SEM from five independent experiments. **p ≤ 0.001. (B) U2-OS cells expressing HA-DYRK1Awt were treated during 8 h with the inhibitor SB216763 (50 μM) or with 0.1% DMSO as vehicle-control. Whole-cell extracts were resolved by 8% SDS-PAGE and immunoblotted with the anti-DYRK1A pS520 antibody (top). The membrane was then stripped and reprobed with anti-HA antibody to detect total HA-DYRK1A protein levels (bottom). (C) U2-OS cells expressing HA-DYRK1Awt were treated with increasing doses of SB216763, and soluble extracts were incubated with recombinant GST-14-3-3β. The amounts of bound DYRK1A (top) and DYRK1A in lysates (10% of inputs) (bottom) in each condition were analyzed by immunoblotting with anti-HA antibody. (D) The chart shows the quantification of the results expressed as the ratio of bound protein to input for each inhibitor concentration. (E) Kinase activities were determined as described in A for HA-DYRK1Awt and HA-DYRK1A/S520A expressed in U2-OS cells treated with 50 μM SB216763 for 8 h. For nontreated cells DMSO was used as vehicle-control. The data represent means ± SEM from three independent experiments. *p ≤ 0.05; **p ≤ 0.001.
We next treated cells expressing HA-DYRK1Awt or HA-DYRK1A/S520A with SB216763 and determined the kinase activity in HA-immunocomplexes. Inhibition by SB216763 is competitive and reversible, so it did not interfere with the kinase assay since activity was determined after extensive washing of the anti-HA immunocomplexes to remove the inhibitor and in the presence of excess ATP. The activity of DYRK1Awt was reduced by as much as 50% by this treatment, which is similar to the activity shown by DYRK1A/S520A (Figure 5E). However, the intrinsically lower activity of DYRK1A/S520 was not affected by SB216763. In summary, loss of Ser-520 phosphorylation correlates with both dissociation of 14-3-3β and lower DYRK1A kinase activity.
14-3-3β Binding Increases the Catalytic Activity of DYRK1A
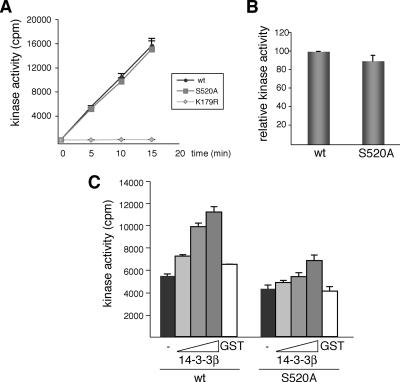
Having established that phosphorylation of Ser-520 in DYRK1A mediates its interaction with 14-3-3β and is required for its catalytic activity in vivo, we wondered whether the effect on DYRK1A activity was caused by the phosphorylation itself or whether it depended on 14-3-3β being bound to DYRK1A through the phospho-Ser residue. To discriminate between these two possibilities, we compared the kinase activity of recombinant GST-fusions of DYRK1Awt and DYRK1A/S520A expressed in bacteria, where DYRK1A Ser-520 is phosphorylated, but there is no 14-3-3. As shown in Figure 6, A and B, both proteins, the wild type and the mutant, have the same specific activity, indicating that the phosphorylation has no effect by itself on the catalytic activity of DYRK1A. However, the addition of purified GST-14-3-3β to the reaction mixture significantly increased DYRK1A activity in a dose-dependent manner, reaching a maximum (2-fold increase) with 1 μg of GST-14-3-3β (Figure 6C). Almost no effect was detected when GST-14-3-3β was added to GST-DYRK1A/S520A; and incubation of DYRK1Awt with GST alone (1 μg) did not affect its catalytic activity. This result strongly implies that the higher in vivo activity of DYRK1Awt compared with the S520A mutant is due to the interaction with 14-3-3β.
Figure 6.
14-3-3β binding to phospho-Ser-520 regulates DYRK1A kinase activity in vitro. (A) Kinase activity of recombinant DYRK1A proteins (GST-DYRK1Awt and the mutants S520A and K179R) was assayed with DYRKtide as substrate. The kinetics of the reaction indicated that 32P incorporation was linear with time and with the enzyme amount used in the assay. (B) The activity of GST-DYRK1Awt versus GST-DYRK1A/S520A is represented as the means ± SEM from three independent experiments, with each point measured in triplicate in the individual experiments. (C) Kinase activities of the indicated GST-DYRK1A proteins were assayed as described in A with increasing amounts of recombinant GST-14-3-3β (0.1, 0.5, and 1 μg) or GST alone (1 μg). The assay was performed three times, giving similar results in each; a representative assay is shown, and the data correspond to the means ± SEM of triplicate measurements.
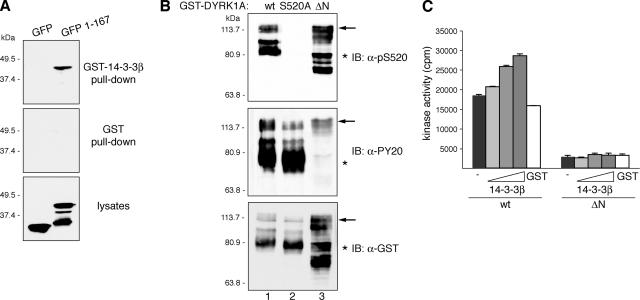
A previous report on 14-3-3 and DYRK1A identified the first 125 amino acids of DYRK1A as the interacting domain, based on the lack of binding of an N-terminal deletion mutant (Kim et al., 2004). To clarify this apparent controversy, we first sought to verify these results, and we found that a GFP-DYRK1A fusion protein comprising the first 167 amino acids was able to bind 14-3-3β in a GST pull-down assay (Figure 7A), confirming that the N terminus does indeed harbor a 14-3-3 binding site. We then checked whether the phosphorylation of Ser-520 was affected by the deletion of the N-terminal region of DYRK1A, as a means of assessing the independence of the two sites. An N-terminally truncated GST-DYRK1A recombinant protein (GST-DYRK1A/ΔN) expressed in bacteria showed similar levels of phosphorylation on Ser-520 as full-length GST-DYRK1A (Figure 7B), indicating that the deletion does not impact on the phosphorylation status of Ser-520 residue. Finally, we also determined the in vitro kinase activity of GST-DYRK1A/ΔN, alone or in the presence of increasing amounts of GST-14-3-3β. The mutant showed significantly lower activity than DYRK1Awt toward DYRKtide (Figure 7C), as reported previously (Himpel et al., 2001). Moreover, we could not detect any increment in the activity of GST-DYRK1A/ΔN when 14-3-3β was added to the reaction (Figure 7C), consistent with a lack of interaction with 14-3-3β. We therefore conclude that the interaction with 14-3-3β is abolished either by the deletion of the N terminus (as reported by Kim et al., 2004) or by the mutation of Ser-520 to an unphosphorylatable residue (this study), suggesting that both sites are necessary to maintain a stable complex between DYRK1A and the 14-3-3 dimer.
Figure 7.
DYRK1A harbors a second 14-3-3 binding site at the N terminus. (A) U2-OS cells were transfected with pEGFP (GFP) or pEGFP-DYRK1A/1-167 (1-167), and lysates were incubated with unfused GST or GST-14-3-3β. Samples were analyzed by immunoblotting with anti-GFP antibody. Lysates represent 10% of inputs. The positions of marker proteins (in kilodaltons) are indicated. (B) Bacterial fusion proteins corresponding to wild-type DYRK1A (wt; lane 1), the mutant DYRK1A/S520A (lane 2), and N-terminally truncated DYRK1A (ΔN; lane 3) were immunoblotted with anti-DYRK1A pS520 (top), anti-phospho-Tyr (PY20; middle), and anti-GST (bottom) antibodies. The positions of marker proteins (in kilodaltons) are indicated. Arrows indicate full-length GST-fusion proteins, and asterisks indicate truncated GST-fusion proteins lacking the C terminus. The abnormal gel mobility of the N-terminal–deleted mutant is explained by its hyperphosphorylation status as shown in Supplemental Figure S6. (C) Kinase activities of GST-DYRK1A proteins were assayed as described in Figure 6C, but GST-DYRK1Awt was compared with the N-terminally truncated mutant GST-DYRK1A/ΔN.
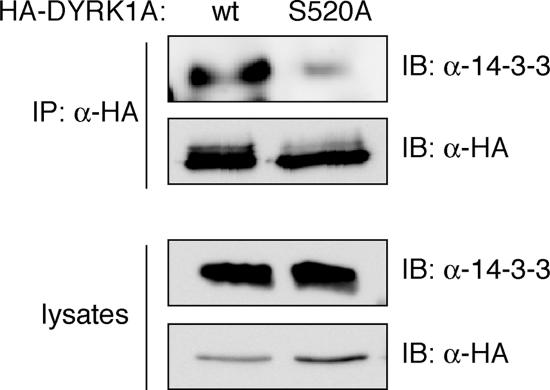
The results from purified proteins were further validated in immunoprecipitation experiments in U2-OS cells transfected with pHA-DYRK1Awt and pHA-DYRK1A/S520A, performed under the same conditions as for the kinase assays where differential kinase activities were detected. Immunoblots of the immunocomplexes with a pan anti-14-3-3 antibody that cross-reacts with most 14-3-3 family members showed that endogenous 14-3-3 proteins coimmunoprecipitate with DYRK1Awt but not with the Ser-520 mutant (Figure 8). All together these findings strongly support a novel mechanism of regulation of DYRK1A activity, in which autophosphorylation of Ser-520 in DYRK1A mediates the formation of a complex with 14-3-3β and modulates the catalytic activity of the kinase.
Figure 8.
14-3-3 binding regulates DYRK1A kinase activity. (A) U2-OS cells expressing HA-DYRK1Awt and the mutant HA-DYRK1A/S520A were immunoprecipitated with anti-HA antibody. The antibody immunoprecipitated similar levels of both HA-tagged DYRK1A proteins, as detected by immunoblot with the same anti- HA antibody. The presence of endogenous 14-3-3 proteins coimmunoprecipitated with DYRK1A was detected by analyzing the samples by immunoblotting with a pan-14-3-3 antibody (top). Cells lysates represented 2% of inputs (bottom). IP, immunoprecipitation.
DISCUSSION
By point mutation analysis and the use of a phospho-specific antibody, we have shown that the binding of DYRK1A to 14-3-3β requires the phosphorylation of Ser-520, a residue located in the C terminus of the protein and framed in a 14-3-3-consensus mode I motif. The association of DYRK1A and 14-3-3 had been reported previously (Kim et al., 2004), but the authors did not identify any requirement for phosphorylation for the interaction. One explanation for this apparently conflicting result might be the differential resistance to phosphatase treatment of phosphorylated Ser-520 and the phosphorylated Tyr residues that were used by Kim et al. (2004) as the control for their phosphatase treatment (Supplemental Figure S3, C). Another possible explanation could lie on the existence of different experimental conditions or 14-3-3 isotype specificity. Nonetheless, all the experimental evidence provided in the present study strongly suggests that the interaction between DYRK1A and 14-3-3β is phosphorylation dependent, as is the case for most 14-3-3 partners (Aitken, 2006).
In addition to the phospho-Ser-520 binding site, there is a second 14-3-3 interacting domain in DYRK1A, within the first 150 amino acids of the N terminus. Both sites seem to be necessary for 14-3-3 binding, because mutation of either greatly diminishes the interaction. The presence of two 14-3-3 binding sites on a single polypeptide increases 14-3-3 binding affinity by more than 30-fold (Yaffe et al., 1997), and several 14-3-3 targets, such as DAF-16 (Cahill et al., 2001) and Cdc25B (Giles et al., 2003), have been reported to require the presence of more than one binding site for stable 14-3-3 association. Given the dimeric nature of 14-3-3, the possibility exists that a molecule of DYRK1A binds to a 14-3-3 dimer through contacts between the phospho-Ser-520 motif and one 14-3-3 monomer and between the second binding motif at the N terminus and the other monomer. Based on the fact that the DYRK1A N-terminal region is able to bind to 14-3-3 when fused to a heterologous protein, but not in the context of the Ser-520 mutation, we propose that 14-3-3 could first recognize and bind to the phospho-Ser-520 motif, which would facilitate binding to the other 14-3-3 binding site in the N terminus, resulting in the stabilization of the complex.
In addition to 14-3-3β, we have found that DYRK1A can also bind 14-3-3η (Supplemental Figure S7). Two recent proteomic analyses have identified DYRK1A as one of the proteins present in complexes of 14-3-3 isoforms γ and σ (Jin et al., 2004; Benzinger et al., 2005); and Kim et al. (2004) have reported interaction with the isoform 14-3-3ε. Although these data would indicate that DYRK1A is able to interact with several 14-3-3 isoforms, further investigation will be needed to determine whether there is any selectivity in DYRK1A/14-3-3 interactions.
The phosphorylation state of some 14-3-3 isoforms, including isoform β, seems to regulate both dimerization and target binding (Yoshida et al., 2005). Thus, we checked whether DYRK1A can act as a putative 14-3-3 kinase. We found no significant phosphorylation of 14-3-3β by in vitro kinase assays (Supplemental Figure S8), suggesting that this is not the case.
The results documented in this manuscript support the idea that DYRK1A phosphorylation at Ser-520 is an intramolecular autophosphorylation event. The phosphorylated Ser residue is not located in an unambiguous DYRK1A consensus site, because there is Asp instead of Pro in the P+1 position; however, the presence of Arg at P−3 would accommodate DYRK1A preferences at that position (Himpel et al., 2000). It is worth noting that the definition of the DYRK1A consensus phosphorylation sequence was based on intermolecular reactions on peptides, and it is conceivable that other sequence requirements are necessary for intramolecular autophosphorylation reactions.
The phosphorylation status of Ser-520 does not interfere with Tyr autophosphorylation, suggesting that the DYRK1A activation loop autophosphorylation is very likely to be an earlier event. Moreover, we can detect phosphorylation of endogenous DYRK1A on Ser-520 in vivo. Thus, apart from the Tyr-321 of the activation loop, Ser-520 represents the only in vivo DYRK1A phosphosite identified to date.
On first consideration, an autophosphorylation event might be assumed to be stoichiometric, but the immunoblot patterns of total DYRK1A and pS520 were not completely equivalent when the kinase was exogenously expressed in mammalian cells and bacteria. Moreover, the levels of phosphorylation of Ser-520 were lower in mammalian cells than in bacterially expressed protein, ranging from 10 to 40% of the recombinant protein. This led us to think that not all DYRK1A molecules were phosphorylated in vivo. This suggestion is supported by the dynamic changes in the level of Ser-520 phosphorylation of endogenous DYRK1A observed in response to bFGF stimulation. Although we cannot exclude the possibility that bFGF signaling affects the autophosphorylation activity itself, it seems more likely that the signaling impacts on the stability of the phosphorylated Ser-520. This might be achieved through the activation of a Ser-520 phosphatase and/or by hampering 14-3-3 binding, which could protect Ser-520 against dephosphorylation, as has been recently reported for Ser-526 in mitogen-activated protein kinase kinase kinase 3 (MEKK3) (Fritz et al., 2006).
A particularly interesting consequence of the interaction between DYRK1A and 14-3-3 is the positive regulation of DYRK1A kinase activity. 14-3-3β seems to increase the catalytic activity of recombinant DYRK1A only toward exogenous substrates, because no effect was detected on DYRK1A autophosphorylation (Supplemental Figure 8). Mechanistically, several scenarios can be envisaged. In one of them, the effect requires binding through both 14-3-3 binding sites, consistent with a model in which the binding of dimeric 14-3-3 to two binding sites on DYRK1A could induce a conformational change in DYRK1A that enhances its enzymatic activity. This mechanism has been proposed for other enzymes that are regulated by 14-3-3 proteins, such as N-acetyltransferase (Obsil et al., 2001). Alternatively, the N-terminal region act as a structural requirement to stabilize the more active conformation, in agreement with the reduced basal catalytic activity of the N-terminal deleted mutant toward exogenous substrates. In this context, DYRK1A would occupy only one of the binding sites in the 14-3-3 dimer, leaving the other available to recruit either DYRK1A substrates or effectors. This has been reported, for example, for one of the 14-3-3 binding sites in Raf proteins, in which 14-3-3 functions as a scaffold mediating the heterooligomerization between the C-Raf and B-Raf kinases (Garnett et al., 2005). In this connection, it is worth mentioning that tau and FKHR, two well-known 14-3-3 targets, are substrates of DYRK1A (Woods et al., 2001).
Only a few of the kinases that bind 14-3-3 have been reported to modulate their activity as a result of this interaction (Table 1). In all cases, 14-3-3 binding depends on a phosphorylated motif. The phosphorylated residue can occur within or outside the catalytic domain, and 14-3-3 binding can lead either to inhibition or to activation of the kinase activity. Although in some cases phosphorylation in 14-3-3 binding motifs is mediated by an upstream kinase, there are three other kinases, in addition to DYRK1A, in which binding to 14-3-3 is known to be regulated by autophosphorylation. In the cases of MEKK3 (Fritz et al., 2006) and 3-phosphoinositide–dependent protein kinase-1 (Sato et al., 2002), catalytic activity is altered by 14-3-3 binding to an autophosphorylated residue within the activation loop. For protein kinase C (PKC)μ, 14-3-3 binding within the regulatory C1 domain inhibits PKC kinase activity (Hausser et al., 1999). Thus, to our knowledge DYRK1A represents the only case in which autophosphorylation of a site outside the catalytic domain leads to an enhanced kinase activity upon 14-3-3 binding. Moreover, this mechanism seems to be specific for DYRK1A, because the Ser-520 motif is not conserved in other members of the DYRK family.
Table 1.
Effect of 14-3-3 binding on the activity of mammalian protein kinases
| Kinase | Binding site | Upstream kinase | Activitya | Reference |
|---|---|---|---|---|
| PI3K | n.d. | n.d. | − | Bonnefoy-Berard et al. (1995) |
| PKCμ | RRLpSNV (Ser-205) | Autob | − | Hausser et al. (1999) |
| RTSpSAE (Ser-219) | ||||
| Raf-1 | RSApSEP (Ser-621) | n.d. | + | Yip-Schneider et al. (2000) |
| RSTpSTP (Ser-259) | AKT, PKA | − | Light et al. (2002) | |
| TESK1 | RCRpSLP (Ser-437) | n.d. | − | Toshima et al. (2001) |
| Wee1 | RSVpSLT (Ser-642) | Chk1 | + | Rothblum-Oviatt et al. (2001) |
| PDK1 | RANpSFV (Ser-241, catc) | Auto | − | Sato et al. (2002) |
| RSK1 | TRLpSKE (Ser-154, cat) | n.d. | − | Cavet et al. (2003) |
| BMK1 | LLKpSLR (Ser-486) | n.d. | − | Zheng et al. (2004) |
| ASK1 | RSIpSLP (Ser-967, cat) | n.d. | − | Goldman et al. (2004) |
| CaMKK1 | RKFpSLQ (Ser-74) | PKA | − | Davare et al. (2004) |
| GSK-3β | RTTpSFA (Ser-9) | AKT | + | Yuan et al. (2004) |
| SIK | PLSpTWC (Thr-182, cat) | LKB | + | Al-Hakim et al. (2005) |
| MEKK3 | GIRpSVT (Ser-526, cat) | Auto | + | Fritz et al. (2006) |
| PKB/AKT | n.d. | n.d. | − | Yang et al. (2006) |
| DYRK1A | RARpSDP (Ser-520) | Auto | + | This study |
n.d., not described.
a +, stimulation of kinase activity; −, inhibition of kinase activity.
b Auto, autophosphorylation.
c cat, phosphorylated residue is located within the catalytic domain.
In summary, our results suggest a model in which the catalytic activity of DYRK1A is regulated by autophosphorylation and binding to 14-3-3β protein. In this model, phosphorylation on Ser-520, outside the DYRK1A catalytic domain, would trigger the association with 14-3-3β. This interaction would induce a conformational change, resulting in increased DYRK1A catalytic activity. At first glance, the twofold increase in kinase activity induced by 14-3-3 binding might seem to be not very impressive. However, one should not forget that Dyrk1A heterozygous mice, with a 50% reduction in expression at the protein level, exhibit clear phenotypic alterations (Fotaki et al., 2002). This, together with the alterations reported in low-overexpressing transgenic mice (Smith et al., 1997; Altafaj et al., 2001; Arron et al., 2006), strongly supports the idea of DYRK1A being a highly gene dosage-sensitive gene, and it allows us to predict that small variations in activity could result in measurable changes in defined biological outputs. Finally, this work argues against the current view of DYRK1A as a constitutive kinase that acquires full competency during its translation and provides the first clear evidence of a mechanism of regulation for DYRK1A activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alicia Raya for technical assistance, M. L. Arbonés (CRG, Barcelona, Spain) for the mouse tissues, and Simon Bartlett for English editorial work. This research was supported by Spanish Ministry of Education and Science grant BFU2004-01768. M.A. was a fellow of the Spanish Ministry of Health (BEFI Program). S.A. is a FI fellow (Generalitat de Catalunya). We also acknowledge the support of the CIBER en Enfermedades Raras.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0668) on January 17, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Aitken A. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Al-Hakim A. K., Goransson O., Deak M., Toth R., Campbell D. G., Morrice N. A., Prescott A. R., Alessi D. R. 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J. Cell Sci. 2005;118:5661–5673. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- Altafaj X., et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum. Mol. Genet. 2001;10:1915–1923. doi: 10.1093/hmg/10.18.1915. [DOI] [PubMed] [Google Scholar]
- Alvarez M., Estivill X., de la Luna S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 2003;116:3099–3107. doi: 10.1242/jcs.00618. [DOI] [PubMed] [Google Scholar]
- Arron J. R., et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Becker W., Joost H. G. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- Benzinger A., Muster N., Koch H. B., Yates J. R., 3rd, Hermeking H. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol. Cell Proteomics. 2005;4:785–795. doi: 10.1074/mcp.M500021-MCP200. [DOI] [PubMed] [Google Scholar]
- Bonnefoy-Berard N., Liu Y. C., von Willebrand M., Sung A., Elly C., Mustelin T., Yoshida H., Ishizaka K., Altman A. Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3-3 proteins in T cells. Proc. Natl. Acad. Sci. USA. 1995;92:10142–10146. doi: 10.1073/pnas.92.22.10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill C. M., Tzivion G., Nasrin N., Ogg S., Dore J., Ruvkun G., Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J. Biol. Chem. 2001;276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- Cavet M. E., Lehoux S., Berk B. C. 14-3-3beta is a p90 ribosomal S6 kinase (RSK) isoform 1-binding protein that negatively regulates RSK kinase activity. J. Biol. Chem. 2003;278:18376–18383. doi: 10.1074/jbc.M208475200. [DOI] [PubMed] [Google Scholar]
- Chen-Hwang M. C., Chen H. R., Elzinga M., Hwang Y. W. Dynamin is a minibrain kinase/dual specificity Yak1-related kinase 1A substrate. J. Biol. Chem. 2002;277:17597–17604. doi: 10.1074/jbc.M111101200. [DOI] [PubMed] [Google Scholar]
- Cohen P., Frame S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Davare M. A., Saneyoshi T., Guire E. S., Nygaard S. C., Soderling T. R. Inhibition of calcium/calmodulin-dependent protein kinase kinase by protein 14-3-3. J. Biol. Chem. 2004;279:52191–52199. doi: 10.1074/jbc.M409873200. [DOI] [PubMed] [Google Scholar]
- de Graaf K., Czajkowska H., Rottmann S., Packman L. C., Lilischkis R., Luscher B., Becker W. The protein kinase DYRK1A phosphorylates the splicing factor SF3b1/SAP155 at Thr434, a novel in vivo phosphorylation site. BMC Biochem. 2006;7:7. doi: 10.1186/1471-2091-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf K., Hekerman P., Spelten O., Herrmann A., Packman L. C., Bussow K., Muller-Newen G., Becker W. Characterization of cyclin L2, a novel cyclin with an arginine/serine-rich domain: phosphorylation by DYRK1A and colocalization with splicing factors. J. Biol. Chem. 2004;279:4612–4624. doi: 10.1074/jbc.M310794200. [DOI] [PubMed] [Google Scholar]
- Dougherty M. K., Morrison D. K. Unlocking the code of 14-3-3. J. Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- Fotaki V., et al. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol. Cell Biol. 2002;22:6636–6647. doi: 10.1128/MCB.22.18.6636-6647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz A., Brayer K. J., McCormick N., Adams D. G., Wadzinski B. E., Vaillancourt R. R. Phosphorylation of serine 526 is required for MEKK3 activity, and association with 14-3-3 blocks dephosphorylation. J. Biol. Chem. 2006;281:6236–6245. doi: 10.1074/jbc.M509249200. [DOI] [PubMed] [Google Scholar]
- Ganguly S., Weller J. L., Ho A., Chemineau P., Malpaux B., Klein D. C. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. USA. 2005;102:1222–1227. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M. J., Rana S., Paterson H., Barford D., Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Giles N., Forrest A., Gabrielli B. 14-3-3 acts as an intramolecular bridge to regulate cdc25B localization and activity. J. Biol. Chem. 2003;278:28580–28587. doi: 10.1074/jbc.M304027200. [DOI] [PubMed] [Google Scholar]
- Goldman E. H., Chen L., Fu H. Activation of apoptosis signal-regulating kinase 1 by reactive oxygen species through dephosphorylation at serine 967 and 14-3-3 dissociation. J. Biol. Chem. 2004;279:10442–10449. doi: 10.1074/jbc.M311129200. [DOI] [PubMed] [Google Scholar]
- Guimera J., Casas C., Estivill X., Pritchard M. Human minibrain homologue (MNBH/DYRK1): characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics. 1999;57:407–418. doi: 10.1006/geno.1999.5775. [DOI] [PubMed] [Google Scholar]
- Gwack Y., et al. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- Hammerle B., Vera-Samper E., Speicher S., Arencibia R., Martinez S., Tejedor F. J. Mnb/Dyrk1A is transiently expressed and asymmetrically segregated in neural progenitor cells at the transition to neurogenic divisions. Dev. Biol. 2002;246:259–273. doi: 10.1006/dbio.2002.0675. [DOI] [PubMed] [Google Scholar]
- Hausser A., Storz P., Link G., Stoll H., Liu Y. C., Altman A., Pfizenmaier K., Johannes F. J. Protein kinase C mu is negatively regulated by 14-3-3 signal transduction proteins. J. Biol. Chem. 1999;274:9258–9264. doi: 10.1074/jbc.274.14.9258. [DOI] [PubMed] [Google Scholar]
- Himpel S., et al. Identification of the autophosphorylation sites and characterization of their effects in the protein kinase DYRK1A. Biochem. J. 2001;359:497–505. doi: 10.1042/0264-6021:3590497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himpel S., Tegge W., Frank R., Leder S., Joost H. G., Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- Jin J., et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Jones D. H., Ley S., Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368:55–58. doi: 10.1016/0014-5793(95)00598-4. [DOI] [PubMed] [Google Scholar]
- Kelly P. A., Rahmani Z. DYRK1A enhances the mitogen-activated protein kinase cascade in PC12 cells by forming a complex with Ras, B-Raf, and MEK1. Mol. Biol. Cell. 2005;16:3562–3573. doi: 10.1091/mbc.E04-12-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., et al. Regulation of Dyrk1A kinase activity by 14-3-3. Biochem. Biophys. Res. Commun. 2004;323:499–504. doi: 10.1016/j.bbrc.2004.08.102. [DOI] [PubMed] [Google Scholar]
- Li K., Zhao S., Karur V., Wojchowski D. M. DYRK3 activation, engagement of protein kinase A/cAMP response element-binding protein, and modulation of progenitor cell survival. J. Biol. Chem. 2002;277:47052–47060. doi: 10.1074/jbc.M205374200. [DOI] [PubMed] [Google Scholar]
- Light Y., Paterson H., Marais R. 14-3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol. Cell Biol. 2002;22:4984–4996. doi: 10.1128/MCB.22.14.4984-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochhead P. A., Sibbet G., Kinstrie R., Cleghon T., Rylatt M., Morrison D. K., Cleghon V. dDYRK2: a novel dual-specificity tyrosine-phosphorylation-regulated kinase in Drosophila. Biochem. J. 2003;374:381–391. doi: 10.1042/BJ20030500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochhead P. A., Sibbet G., Morrice N., Cleghon V. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell. 2005;121:925–936. doi: 10.1016/j.cell.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Mao J., Maye P., Kogerman P., Tejedor F. J., Toftgard R., Xie W., Wu G., Wu D. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- Marti E., Altafaj X., Dierssen M., de la Luna S., Fotaki V., Alvarez M., Perez-Riba M., Ferrer I., Estivill X. Dyrk1A. expression pattern supports specific roles of this kinase in the adult central nervous system. Brain Res. 2003;964:250–263. doi: 10.1016/s0006-8993(02)04069-6. [DOI] [PubMed] [Google Scholar]
- Moriya H., Shimizu-Yoshida Y., Omori A., Iwashita S., Katoh M., Sakai A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 2001;15:1217–1228. doi: 10.1101/gad.884001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. T., et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsil T., Ghirlando R., Klein D. C., Ganguly S., Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell. 2001;105:257–267. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- Okui M., Ide T., Morita K., Funakoshi E., Ito F., Ogita K., Yoneda Y., Kudoh J., Shimizu N. High-level expression of the Mnb/Dyrk1A gene in brain and heart during rat early development. Genomics. 1999;62:165–171. doi: 10.1006/geno.1999.5998. [DOI] [PubMed] [Google Scholar]
- Rothblum-Oviatt C. J., Ryan C. E., Piwnica-Worms H. 14-3-3 binding regulates catalytic activity of human Wee1 kinase. Cell Growth Differ. 2001;12:581–589. [PubMed] [Google Scholar]
- Sato S., Fujita N., Tsuruo T. Regulation of kinase activity of 3-phosphoinositide-dependent protein kinase-1 by binding to 14-3-3. J. Biol. Chem. 2002;277:39360–39367. doi: 10.1074/jbc.M205141200. [DOI] [PubMed] [Google Scholar]
- Skurat A. V., Dietrich A. D. Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J. Biol. Chem. 2004;279:2490–2498. doi: 10.1074/jbc.M301769200. [DOI] [PubMed] [Google Scholar]
- Smith D. J., et al. Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat. Genet. 1997;16:28–36. doi: 10.1038/ng0597-28. [DOI] [PubMed] [Google Scholar]
- Toshima J. Y., Toshima J., Watanabe T., Mizuno K. Binding of 14-3-3beta regulates the kinase activity and subcellular localization of testicular protein kinase 1. J. Biol. Chem. 2001;276:43471–43481. doi: 10.1074/jbc.M104620200. [DOI] [PubMed] [Google Scholar]
- Wegiel J., et al. Cell type- and brain structure-specific patterns of distribution of minibrain kinase in human brain. Brain Res. 2004;1010:69–80. doi: 10.1016/j.brainres.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Woods Y. L., Rena G., Morrice N., Barthel A., Becker W., Guo S., Unterman T. G., Cohen P. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 2001;355:597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. The structural basis for 14-3-3, phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Yang E. J., Ahn Y. S., Chung K. C. Protein kinase Dyrk1 activates cAMP response element-binding protein during neuronal differentiation in hippocampal progenitor cells. J. Biol. Chem. 2001;276:39819–39824. doi: 10.1074/jbc.M104091200. [DOI] [PubMed] [Google Scholar]
- Yang H., et al. DNA damage-induced protein 14-3-3 sigma inhibits protein kinase B/Akt activation and suppresses Akt-activated cancer. Cancer Res. 2006;66:3096–3105. doi: 10.1158/0008-5472.CAN-05-3620. [DOI] [PubMed] [Google Scholar]
- Yip-Schneider M. T., Miao W., Lin A., Barnard D. S., Tzivion G., Marshall M. S. Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem. J. 2000;351:151–159. doi: 10.1042/0264-6021:3510151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Yamaguchi T., Natsume T., Kufe D., Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat. Cell Biol. 2005;7:278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
- Yuan Z., Agarwal-Mawal A., Paudel H. K. 14-3-3 binds to and mediates phosphorylation of microtubule-associated tau protein by Ser9-phosphorylated glycogen synthase kinase 3beta in the brain. J. Biol. Chem. 2004;279:26105–26114. doi: 10.1074/jbc.M308298200. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Yin G., Yan C., Cavet M., Berk B. C. 14-3-3beta binds to big mitogen-activated protein kinase 1 (BMK1/ERK5) and regulates BMK1 function. J. Biol. Chem. 2004;279:8787–8791. doi: 10.1074/jbc.M310212200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.