Abstract
Eukaryotic nuclei contain three classes of multisubunit DNA-directed RNA polymerase. At the core of each complex is a set of 12 highly conserved subunits of which five—RPB5, RPB6, RPB8, RPB10, and RPB12—are thought to be common to all three polymerase classes. Here, we show that four distantly related eukaryotic lineages (the higher plant and three protistan) have independently expanded their repertoire of RPB5 and RPB6 subunits. Using the protozoan parasite Trypanosoma brucei as a model organism, we demonstrate that these distinct RPB5 and RPB6 subunits localize to discrete subnuclear compartments and form part of different polymerase complexes. We further show that RNA interference-mediated depletion of these discrete subunits abolishes class-specific transcription and hence demonstrates complex specialization and diversification of function by conventionally shared subunit groups.
INTRODUCTION
DNA-directed RNA polymerases form the crux of a highly dynamic process fundamental to biology. In eukaryotic nuclei, the majority of RNA synthesis is mediated by three multisubunit RNA polymerase complexes (Roeder and Rutter, 1969), each of which is independently regulated and is responsible for transcribing a nonoverlapping subset of genes (Roeder et al., 1976). As a general consensus, DNA-directed RNA polymerase (Pol) I transcribes rRNA genes, Pol II transcribes protein coding genes and some small nuclear/nucleolar RNAs, and Pol III transcribes tRNA genes and other small nuclear RNAs.
Most of what is known about the composition of RNA polymerase complexes originates from studies on fungi (Sentenac, 1985) and from sequence and functional homology inferred between fungi and metazoans (Shpakovski et al., 1995). Each of the three multisubunit polymerases is composed of a core set of 12 highly conserved subunits. Of this core set, some subunits have unique paralogues for each polymerase class and others are shared by Pol I and Pol III or by all three polymerases. In yeast and metazoans, five subunits (RPB5, RPB6, RPB8, RPB10, and RPB12) are common to all three classes of RNA polymerase and are conventionally referred to as common or shared subunits (Buhler et al., 1980; Lanzendorfer et al., 1997). Alongside the core set, each polymerase complex also contains a number of specific subunits that are neither common nor have paralogous counterparts in the other polymerase classes. This is particularly evident for Pol III in yeast, where subunits RPC82, RPC53, RPC37, RPC34, and RPC31 do not have paralogous counterparts in either Pol I or Pol II complexes. Previous reports have identified expansions within the shared subunit repertoire in the genomes of three distantly related eukaryotes, Arabidopsis thaliana (Larkin et al., 1999), Giardia lamblia (Best et al., 2004), and three trypanosomatids (Trypanosoma brucei, T. cruzi, and Leishmania major) (Kelly et al., 2005; Walgraffe et al., 2005; Devaux et al., 2006). However, the function of these putative subunits has not been addressed.
Trypanosomatids are the causative agents of several globally important parasitic diseases, and as such they are the focus of much scientific attention. The African trypanosome T. brucei is radical among eukaryotes, because it has evolved a multifunctional Pol I complex capable of transcribing both rRNA (as in other organisms) and protein coding genes (Gunzl et al., 2003). The repertoire of protein-coding genes transcribed by this polymerase is limited to those encoding the major surface coat proteins (and associated genes) of its different life cycle stages. Intriguingly, in bloodstream-form cells, these two modes of Pol I transcription are spatially separated within the nucleus. rRNAs are transcribed in the primary Pol I-containing organelle, the nucleolus. However, genes encoding the bloodstream-form surface coat proteins are transcribed by Pol I in an extranucleolar expression site body (ESB) (Navarro and Gull, 2001). This is the only documented example of endogenous protein-coding gene transcription by Pol I; thus, it presents the trypanosome with the challenge of coupling mRNA processing to Pol I transcription.
The trypanosomatid transcriptional complexity is not only limited to Pol I but also extends to Pol II. In T. brucei and other members of the trypanosomatid family, Pol II transcription occurs in the absence of detectable promoters (Vanhamme and Pays, 1995; Clayton, 2002). Unlike other eukaryotes, mRNA capping is mediated by trans-splicing of a short, capped sequence to the 5′ end of all mRNAs (LeBowitz et al., 1993; Ullu et al., 1993). This process uncouples Pol II transcription initiation from mRNA capping and is one factor that facilitates the transcription of all protein coding genes in long polycistronic units. Astonishingly, this uncoupled polycistronic transcription is achieved in the absence of many basal transcription factors otherwise conserved throughout Archaea and Eukarya (Ivens et al., 2005; Palenchar and Bellofatto, 2006).
Our previous bioinformatic analysis has shown that the full complement of Pol II subunits, including those that are shared with Pol I and Pol III, are present in T. brucei (Kelly et al., 2005). However, some of the smaller Pol I and Pol III subunits could not be detected. Intriguingly, of the five shared subunits, both RPB5 and RPB6 have two paralogues in T. brucei. This is also true for T. cruzi and L. major, two related trypanosomatids for which genomic data are available. Tandem affinity purification identified an association between one of the two RPB5 isoforms and the Pol I subunit TbRPA12 in T. brucei (Walgraffe et al., 2005), whereas the other TbRPB5 was found to associate with the Pol II subunit TbRPB9 (Devaux et al., 2006). Similarly, an unstable association between epitope-tagged RPB6z and the largest subunit of Pol I was also detected (Nguyen et al., 2006). Here, we use a number of approaches to demonstrate for the first time diversification of function and complex specialization by different isoforms of the conventionally shared RNA polymerase subunits RPB5 and RPB6.
MATERIALS AND METHODS
Subunit Identification and Bioinformatic Analysis
Iterative profile-based searching techniques are a powerful way of identifying homologues in diverse, distantly related organisms. To produce seed alignments for such techniques, simple BLAST searches (Altschul et al., 1997) by using the Saccharomyces cerevisiae Rpb5p (NP_009712) or Rpb6p (NP_015513) were performed. Several sequences with low expectation values from closely related organisms were easily identified. These sequences were aligned using ClustalX (Thompson et al., 1994) and manually edited before being used to generate hidden Markov models (Eddy, 1998). These models were then used to search a large, protein sequence database from a diverse set of organisms (see Supplemental Material). The resultant hits were aligned to the model, and unconvincing low-scoring matches were discarded. A new alignment containing the additional sequence matches was then used to generate another hidden Markov model, and the database was researched. This process was repeated until no further satisfactory matches could be detected.
The final amino-acid alignments were manually edited and trimmed to informative blocks of 188 and 79 characters for RPB5 and RPB6, respectively. These alignments were then used to infer Bayesian maximum likelihood trees by using the program MrBayes 3.0b4 (Huelsenbeck and Ronquist, 2001). The WAG substitution matrix was used with a gamma-distributed substitution rate variation approximated by four discrete categories with shape parameter estimated from the data. Bootstrap support for the inferred topologies was estimated from 100 replicate Bayesian tree inferences with character resampling. One thousand bootstrap tree estimations were also made under maximum parsimony and neighbor-joining schema by using the program PAUP*4.0b10 (Swofford, 1998).
The protein sequence alignments were additionally used to model each subunit by using the SWISS-MODEL homology modeling server (Schwede et al., 2003). RPB5 sequences were modeled in reference to protein chain E from the ExPDB template file 1Y1V, RPB6 in reference to chain F of the same template (Kettenberger et al., 2004).
Cell Culture
Procyclic-form cells were grown at 27°C in SDM-79 medium (Brun and Jenni, 1977) supplemented with either 10 or 15% fetal bovine serum. Bloodstream-form cells were grown at 37°C in 5% CO2 in HMI-9 medium (Hirumi and Hirumi, 1989) supplemented with either 20% fetal bovine serum or 10% fetal bovine serum with 10% serum plus. Cloning of procyclic-form cells was performed by limiting dilution on 96-well plates. Cloning of bloodstream-form cells was performed either by limiting dilution on 24-well plates or by plating cells on agarose (Carruthers and Cross, 1992).
Constructs Generation and Transfection
RNA Interference (RNAi).
Five hundred base-pair fragments of TbRPB5 (XP_827817), TbRPB5z (XP_827816), TbRPB6 (XP_825362, XP_825364), and TbRPB6z (XP_828112) were amplified by polymerase chain reaction (PCR) by using specific primers and inserted into the inducible double-stranded RNA (dsRNA)-expression vectors pZJM (Wang et al., 2000) or p2T7-177 (Wickstead et al., 2002) by using appropriate sites. These vectors were linearized with NotI restriction endonuclease and then transfected into procyclic form 29-13 cells (Wirtz et al., 1999) or bloodstream form “single-marker” cells (Wirtz et al., 1999) as described previously (Wirtz et al., 1999). Stable transformants were selected with 5 or 1 μg ml−1 phleomycin, respectively. After selection, cells lines were removed from phleomycin for 48 h before induction of RNAi mediated by addition of 1 μg ml−1 doxycycline (Sigma-Aldrich, St. Louis, MO) to culture medium.
Chimera Expression.
To generate cell lines expressing (from their endogenous loci) N-terminal enhanced green fluorescent protein (GFP) chimeras of each RPB5 and RPB6 subunit, ∼250 base-pair fragments were amplified by PCR from the 5' end of each gene and from the 3′ end of the respective upstream intergenic region by using primers incorporating suitable restriction sites. These fragments were simultaneously ligated as open reading frame/intergenic region pairs into the pEnT5 vector (Kelly, Reed, Kramer, Ellis, Webb, Sunter, Salje, Marinsek, Wickstead, Gull, and Carrington, unpublished data) by using the XbaI and BamHI sites. The vector was linearized by XhoI before transfection into procyclic form Lister 427 cells or bloodstream form “single-marker” cells (Wirtz et al., 1999) as stated above. Stable transformants were selected with 50 and 5 μg ml−1 hygromycin (Sigma-Aldrich), respectively.
Tandem Affinity Purification (TAP) Tagging.
The open reading frames encoding TbRPB5 and TbRPB5z were amplified by PCR from T. brucei genomic DNA and inserted into the pLew100 NLS-NTAP tag vector (Devaux et al., 2006) by using the XhoI and XbaI sites. After stable transfection into 29–13 cells (Wirtz et al., 1999), the expression of TAP-tagged proteins was induced by addition of 1 μg ml−1 doxycycline to culture medium.
Immunofluorescence
Cells were fixed for 5 min in 2% paraformaldehyde by addition of the fixative directly to the culture medium. They were subsequently washed twice in phosphate-buffered saline (PBS; 137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4) and allowed to settle onto glutaraldehyde-derivatized 3-aminopropyltriethoxysilane-coated (Sigma-Aldrich) slides. Cells were then permeabilized with 0.1% NP-40, 0.2% arginine in PBS and washed with 0.2% arginine in PBS. Cells were subsequently incubated with rabbit polyclonal anti-GFP (gift from Jeff Errington, Newcastle University, United Kingdom) and mouse monoclonal L1C6 primary antibodies, washed, and then incubated with goat anti-rabbit AlexaFluor 488 (Invitrogen, Carlsbad, CA) and goat anti-mouse tetramethylrhodamine B isothiocyanate (Sigma-Aldrich)-conjugated secondary antibodies. Slides were washed and mounted in Vectashield anti-fade fluorescence mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). L1C6 is a monoclonal antibody (mAb) with reactivity to the nucleolus; the precise antigen is unknown.
Tandem Affinity Purification and Protein Identification
Approximately 1010 cells expressing either the TAP-tagged TbRPB5 or TbRPB5z protein were collected by centrifugation and washed in PBS. Cells were resuspended in ice-cold lysis buffer (0.1 M KCl, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 17% glycerol, 0.5% NP-40, and 10 mM Tris, pH 8.0) with protease inhibitors and incubated for 30 min at 4°C with gentle agitation. The supernatants resulting from two successive centrifugation steps at 4°C (10 min at 3000 × g followed by 2 h at 100,000 × g) were adjusted to 150 mM NaCl before using them for standard TAP tag purification (Puig et al., 2001; Devaux et al., 2006). Eluted proteins were acetone precipitated and resuspended in Laemmli buffer before loading on 4–12% Bis-Tris gradient gel (NuPAGE; Invitrogen). The gels were stained with Sypro Ruby reagent (Invitrogen), and the bands were excised from the gel before in-gel tryptic digestion and analysis by mass spectrometry (Devaux et al., 2006).
Analysis of Transcripts
Northern Blot.
Total RNA was extracted by TRIzol reagent (Invitrogen) and analyzed by standard methods.
In Situ Nascent RNA Labeling.
Cells were harvested and washed twice with transcription buffer (100 mM KCl, 5 mM MgCl2, 5 mM DTT, and 0.5 mM EGTA). Cells were subsequently permeabilized with 250 μg ml−1 saponin before incubation in transcription buffer containing ATP, CTP, GTP, and Br-UTP for 15 min at 27°C, with or without the addition of 100 μg ml−1 α-amanitin. Cells were then fixed with 3.5% formaldehyde in PBS and Br-UTP incorporation assayed by immunofluorescence with anti-Br-dUTP antibodies.
Run On Assay.
Run on transcription in permeabilized trypanosomes was performed as described previously (Ullu and Tschudi, 1990). Total labeled RNA was then extracted using the TRIizol reagent (Invitrogen) and quantified by absorbance at 260 nm. Total incorporation was measured by counting after precipitation in 10% trichloroacetic acid. PCR products encompassing the whole open reading frame of each gene was used as hybridization target.
Electron Microscopy
Cells were fixed for 15 min at room temperature in 4% formaldehyde, 0.1% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2. Cell pellets, wrapped in 1% agarose, were dehydrated in increasing concentrations of ethanol and embedded in EMbed 12 (Electron Microscopy Sciences, Hatfield, PA). After polymerization at 60°C for 3 d, samples were cut and ultrathin sections were collected in Formvar-coated nickel grids that were stained with uranyl acetate and lead citrate. Observations were made on a Tecnai 10 electron microscope (FEI, Hillsboro, OR), and images were captured and processed with a MegaView II camera and AnalySIS software (Olympus Soft Imaging Solutions, Münster, Germany).
RESULTS
Database Searching Reveals Duplication of Two Common Subunits in Several Lineages
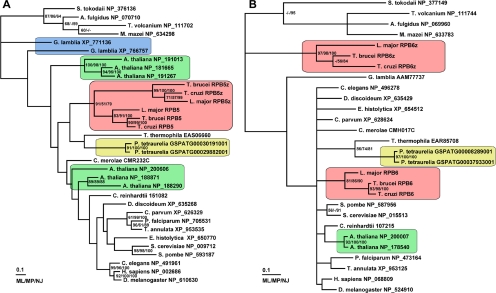
Iterative profile-based searches of 19 disparate eukaryotic genomes resulted in the identification of 29 RPB5 and 24 RPB6 sequences. These predicted protein sequences were aligned and edited, and the alignments were then used to infer Bayesian phylogenies (Huelsenbeck and Ronquist, 2001) for each subunit. The resulting trees (Figure 1A) show that duplication of RPB5 has occurred in multiple eukaryotic lineages. We find that A. thaliana encodes six putative RPB5 proteins that can be divided into two discrete groups of three sequences. The first group (NP_19013, NP_191267, and NP_181665) are characterized by small 12–20 amino acid N-terminal extensions conserved in higher plants. The second group (NP_200606, NP_188871, and NP_188290) lack this short N-terminal extension and include a highly truncated sequence, NP_188290. Multiple RPB5 sequences can be found in the genomes of other higher plants that can similarly be grouped by the presence or absence of this short N-terminal extension (see Supplemental Material). However, only a single RPB5 sequence can be found in the genomes of the red alga Cyanidioschyzon merolae or green alga Chlamydomonas reinhardtii.
Figure 1.
Multiple homologues of the shared subunits RPB5 and RPB6 occur in a broad range of eukaryotes. (A) Bayesian maximum likelihood tree of RPB5 sequences. (B) Bayesian maximum likelihood tree of RPB6 sequences. Instances where multiple paralogues exist are indicated by shaded boxes, GenBank accession numbers are shown with taxon names. Accession numbers for Cyanidioschyzon merolae, Chlamydomonas reinhardtii, and Paramecium tetraurelia proteins are specific for their genome project databases (Matsuzaki et al., 2004, http://www.jgi.doe.gov/ and Arnaiz et al., 2007, respectively). Bootstrap support for the Bayesian topology from maximum likelihood (ML), maximum parsimony (MP), and neighbor-joining (NJ) approaches is indicated next to nodes. Bar, 0.1 changes.
The single-cell eukaryotes G. lamblia, Paramecium tetraurelia, and the three trypanosomatids T. brucei, T. cruzi, and L. major also contain multiple RPB5 sequences. The three trypanosomatids each encode two RPB5 paralogues forming two separate clades, but they are clearly derived from a single common ancestor (Figure 1A). The grouping of these sequences suggests that they arose independently within the trypanosomatid lineage predating the split of L. major from T. cruzi and T. brucei. Similarly for RPB6, it is apparent that duplications have occurred in at least three different eukaryotic lineages. A. thaliana (and other higher plants excluding green and red algae), P. tetraurelia, and the trypanosomatids each encode at least two putative RPB6 proteins in their genomes (Figure 1B).
Variant Subunits Differ from Their Canonical Forms in Similar Ways
The canonical RPB5 subunit is composed of two domains: an N-terminal domain that encompasses ∼60% of the sequence and a smaller C-terminal domain that is homologous to the archaeal RNA polymerase subunit rpoH. The small C-terminal domain is well conserved across Eukarya and archaea and is responsible for the attachment of RPB5 to the largest subunit of the polymerase complex. One trypanosomatid RPB5 homologue (TbRPB5, XP_827817) is identical in domain organization to the canonical eukaryotic RPB5 subunit. The other (TbRPB5z, XP_827816) differs mainly by the presence of three insertions, the two largest of which reside in the eukaryote-specific N-terminal domain (Figure 2A). The third and smallest insertion resides just before the C-terminal domain. Interestingly, an insertion is found at the same position in both paralogues of G. lamblia RPB5 and both paralogues of P. tetraurelia RPB5 (see Supplemental Material). Insertions within the N-terminal region of RPB5 are not specific to trypanosomatids and can also be found in the single putative RPB5 homologue in Tetrahymena thermophila (see Supplemental Material). In trypanosomatids, the N-terminal insertions differ in size and are poorly alignable. However, they share some similarity in their sequence composition, each being composed of a high proportion of charged residues (Figure 2B). These insertions were not used to infer the tree and thus are not responsible for the clustered grouping of the TbRPB5z sequences (Figure 1A).
Figure 2.
Comparison between duplicated subunits in trypanosomatids. For all panels, conserved structure is in blue, and residues that are predicted to cause alterations to the solved structure or residues that are not present in the solved structure are in green. (A) Cartoon depicting domains and insertions in trypanosomatid RPB5 sequences. (B) Comparison between insertion size and proportion of charged residues for the two largest insertions in the three trypanosomatids. (C) Cartoon depicting domains and insertions in trypanosomatid RPB6 sequences. (D) Comparison between the known S. cerevisiae RPB5 structure and those predicted for T. brucei RPB5 and RPB5z. (E) Comparison between the known S. cerevisiae RPB6 structure and those predicted for T. brucei RPB6 and RPB6z. The structure of the RPB6 charged N-terminal domain is not known and hence is not included. (F) Predicted RPB5z and RPB6z subunits placed in context of an RNA polymerase complex (Pol II), black arrow indicates direction and point of entrance for DNA entering the complex.
The level of sequence identity between all the organisms used to infer the RPB5 tree is very low. Of the 188 sequence positions sampled, only four are identical across all sequences in the tree (not including the highly truncated A. thaliana homologue). These correspond to residues R26, H147, D182, and R212 of the S. cerevisiae protein NP_009712. Given this low level of sequence identity, we looked to see how the four residues shown to be important for transcription factor binding in human RPB5 (F76, I104, T111, and S113) (Le et al., 2005) are distributed across other eukaryotes. S113 is the least well conserved of these four residues, only occurring in metazoans and fungi. T111 corresponds to a polar residue in most eukaryotes, excluding the trypanosomatid RPB5z isoforms, Entamoeba histolytica RPB5, both P. tetraurelia homologues, and A. thaliana NP_19103 and NP_188290. I104 is represented by a nonpolar residue in all eukaryotes apart from one G. lamblia isoform, and F76 is conserved in all eukaryotes except for two isoforms of A. thaliana RPB5 (NP_200606 and NP_188290) and the two G. lamblia isoforms (for alignment, see Supplemental Material).
The canonical RPB6 subunit is also composed of two domains: a short negatively charged N-terminal domain and a highly conserved C-terminal domain that is homologous to the archaeal RNA polymerase subunit rpoK. The mean length of the N-terminal domain is 42 amino acids, and it is composed of ∼42% acidic residues. This domain is poorly alignable across the eukaryotes and is absent from archaeal rpoK. Intriguingly, this highly charged N-terminal domain is absent from G. lamblia RPB6 and from the trypanosomatid RPB6 variant RPB6z (Figure 2C), making them seem similar in structure to the archaeal subunit. The trypanosomatid RPB6z subunit also differs from the canonical RPB6 because of a short insertion in the C-terminal domain. A similar insertion is found at the same position in the predicted gene models for RPB6 in the apicomplexan parasites Plasmodium falciparum and Theileria annulata but not in the related alveolates T. thermophila or P. tetraurelia. Unlike the N-terminal insertions of RPB5z, the length and sequence of this small insertion are conserved within the trypanosomatids (for alignment, see Supplemental Material). The level of sequence identity between all the organisms used to infer the RPB6 tree is higher than for RPB5. Of the 79 sequence positions sampled, seven are identical across all taxa. These correspond to residues T86, E89, R92, R97, A98, Q100, and R136 of the S. cerevisiae protein NP_015513.
Novel Insertions Predicted to Form Surface-exposed Structures
To investigate how the insertions found in the noncanonical trypanosomatid RPB5 and RPB6 subunits might affect their interactions with other polymerase components, we used the solved crystal structures for the yeast Pol II complex subunits (Cramer et al., 2000; Todone et al., 2000). The manually evaluated and edited alignments for both RPB5 and RPB6 were used to model the T. brucei subunits with reference to their solved yeast homologues. For T. brucei RPB5z, all three insertions are predicted to form surface-exposed structures (Figure 2D). The most N-terminal proximal insertion is predicted to occupy a region adjacent the DNA entrance channel, creating a novel interaction surface for the polymerase complex facing DNA-bound transcriptional regulators (Figure 2F). The second largest insert is predicted to form a small helix that protrudes away from the core complex (Figure 2, D and F). The third and smallest insertion is predicted to take the form of a small sheet that projects toward RPB6 and the dissociable heterodimer complex.
By homology modeling of T. brucei RPB6z in reference to the solved C-terminal domain of RPB6 (Cramer et al., 2000), we predict an orientation for the sole insertion that disrupts this C-terminal domain in all three trypanosomatids as well as in P. falciparum and T. annulata. This insertion is predicted to form a surface exposed structure that extends across a gap on the side of the Pol II complex (Figure 2, E and F).
The Different Paralogues of RPB5 and RPB6 Localize to Discrete Subnuclear Compartments
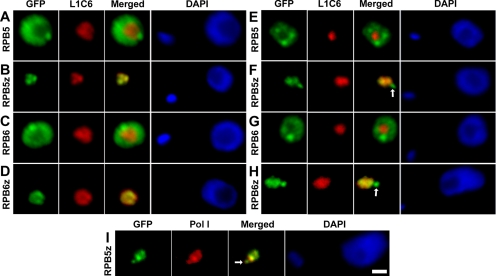
To address whether the multiple RPB5 and RPB6 subunits represent different forms of the same polymerase class or whether they are unique to different polymerase classes, we interrogated their subnuclear localization by generating chimeras of each isoform fused at their N terminus to GFP. Chimeric genes were generated at the endogenous genomic locus for each subunit to preserve, as far as possible, both the endogenous transcriptional environment and the 3′ untranslated region known to be crucial for posttranscriptional regulation in trypanosomatids (Teixeira and daRocha, 2003). In tsetse-form procyclic T. brucei cells, both GFP-TbRPB5 and GFP-TbRPB6 localize to the nucleoplasm excluding the nucleolus (Figure 3, A and C), indicating that they form part of the Pol II and/or Pol III but not Pol I complexes. The signals from both GFP-TbRPB5 and GFP-TbRPB6 seem to extend throughout the nucleoplasm, but they are not uniformly distributed being more concentrated at a few discrete foci. In contrast to the localization observed for GFP-TbRPB5 and GFP-TbRPB6, both GFP-TbRPB5z and GFP-TbRPB6z localize only to the nucleolus—the sole site of Pol I transcription in tsetse-form procyclic cells.
Figure 3.
Subnuclear localization of GFP-tagged RPB5 and RPB6 subunits. (A–D) Tsetse form procyclic cells fixed in 2% paraformaldehyde and labeled with anti-GFP and L1C6 anti-nucleolar antibodies. (E–H) Bloodstream-form cells processed in the same manner. (I) Bloodstream-form cells' colocalization of TbRBP5z-GFP with anti-Pol I largest subunit antibody. ESBs are indicated with white arrows. Bars, 1 μm.
In bloodstream-form cells, the localization patterns for both GFP-TbRPB5 and GFP-TbRPB6 match those observed in tsetse-form cells (Figure 3, E and G). However, both GFP-RPB5z and GFP-RPB6z localize to two discrete subnuclear compartments, the nucleolus and an additional extranucleolar compartment (Figure 3, F and H). This extranucleolar compartment was shown to be the expression site body by colocalization with a mAb to the Pol I largest subunit (Figure 3I; Navarro and Gull, 2001).
The Different Paralogues of RPB5 Form Part of Different Polymerase Complexes
To address whether the different paralogues of RPB5 did indeed form part of different polymerase complexes, we generated procyclic-form T. brucei cells capable of inducibly expressing TAP-tagged chimeras of either TbRPB5 or TbRPB5z. Inducible expression of TAP tag fusion proteins was confirmed by Western blot (Figure 4A) and immunofluorescence. Subnuclear localization of the TAP-tagged proteins agreed with the localization of GFP-TbRPB5 and GFP-TbRPB5z (data not shown). Protein extracts were affinity purified, and the resultant complexes were separated by SDS-PAGE. The highest molecular weight bands of each purified complex were identified by mass spectrometry. Analysis of the resultant complexes revealed that TAP-TbRPB5z copurifies with the two largest Pol I subunits, TbRPA1 and TbRPA2, but no subunits belonging to Pol II or Pol III could be detected (Figure 4B and Supplemental Material). Conversely, affinity purification of TAP-TbRPB5 resulted in copurification of subunits from both Pol II and Pol III, but no subunits belonging to Pol I could be detected (Figure 4B and Supplemental Material).
Figure 4.
TAP-tagged RPB5 subunits interact with different polymerase classes. (A) Expression of the TbRPB5z and TbRPB5 TAP-tagged proteins in procyclic forms. Expression was induced overnight by addition of doxycycline and analyzed by Western blot with anti-TAP tag antibodies. (B) Protein extracts affinity purified from the TbRPB5 or TbRPB5z TAP tag-expressing cell lines were separated on gradient gels and stained by Sypro Ruby. The highest molecular weight bands of each purification were analyzed by mass spectrometry.
RNAi-mediated Depletion of Either TbRPB5 or TbRPB5z Abolishes Class-specific Transcription
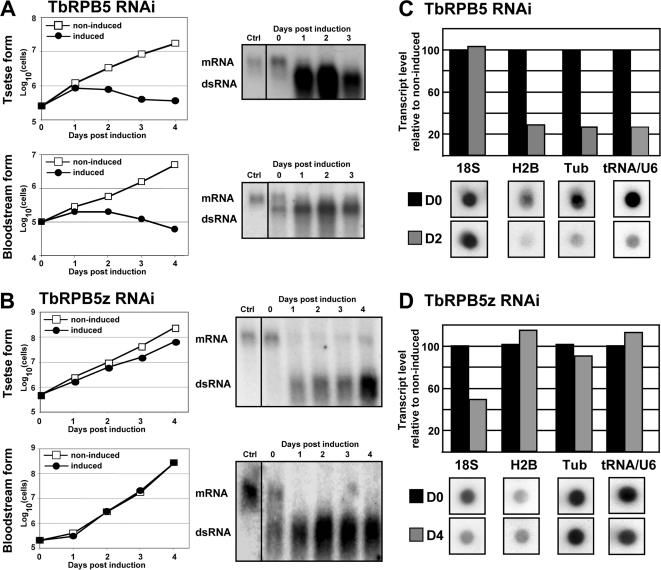
To evaluate the effect of loss of either trypanosome RPB5 subunit on the transcription profile of the cell, tetracycline-inducible RNAi constructs were generated to specifically target degradation of each RPB5 mRNA independently. The level of mRNA and dsRNA was monitored in each case. The induction of dsRNA led to a decrease of mRNA levels of the two genes in both life cycle stages (Figure 5), although TbRPB5z transcripts could still be observed in procyclic forms. Knockdown of TbRPB5 resulted in rapid growth arrest and cell death in both bloodstream- and tsetse-form cells (Figure 5A). Knockdown of TbRPB5z also produced a growth phenotype in tsetse-form cells, but it did not cause any observable growth defect in bloodstream-form cells (Figure 5B). Numerous attempts to generate an RPB5z knockout cell line in both life cycle stages have failed. Similar growth phenotypes were observed for knock-down of TbRPB6 and TbRPB6z in both life cycle stages (see Supplemental Material).
Figure 5.
Effect of RNAi on growth. (A) TbRPB5-RNAi. (B) TbRPB5z-RNAi. In each case, transcript level and production of double-stranded RNA was monitored by Northern blot. (C and D) Dot-blotted DNAs were hybridized with [32P]UTP-labeled RNAs. The dot blotted genes transcribed by RNA polymerase I (18s RNA), II (histone 2B and α-tubulin), and III (tRNA) are indicated. Quantification of hybridization signals is shown in histograms before (D0) or after 2 (D2) or 4 (D4) days of doxycycline induction.
To look at the effect of RNAi on nascent transcription, total cellular [32P]UTP-labeled RNA was isolated from induced cells at either 48 or 96 h postinduction and hybridized to transcript-specific Dot-blotted DNAs (Figure 5). Targeted depletion of TbRPB5z mRNA resulted in reduction in levels of Pol I-transcribed 18s rRNA, but it did not effect levels of histone 2B, α-tubulin, or tRNA-Thr-U6snRNA (Figure 5D). Complementary to this, targeted depletion of TbRPB5 mRNA resulted reduction in levels of Pol II-transcribed genes histone 2B and α-tubulin as well as the Pol III-transcribed tRNA-Thr-U6snRNA, but it had no effect on the Pol I-transcribed 18s RNA (Figure 5C). In agreement with this, targeted depletion of TbRPB5 mRNA resulted in inhibition of nucleoplasmic but not nucleolar RNA synthesis when assessed by Br-UTP incorporation. Conversely, targeted depletion of TbRPB5z mRNA resulted in reduction of α-amanitin resistant (Pol I) RNA synthesis (data not shown).
Targeted Depletion of TbRPB5z Causes Disruption of Nucleolar Structure
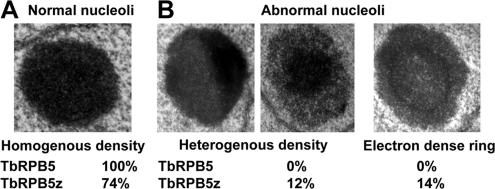
Because the nucleolus is the sole site of Pol I-mediated rRNA transcription in procyclic-form cells, we used transmission electron microscopy to determine whether the knockdown of the Pol I-specific subunit affected nucleolar structure. Both induced and noninduced procyclic-form TbRPB5z and TbRPB5 RNAi cells were fixed and embedded. In each case, 200 induced and 200 control cells were analyzed from thin sections of nuclei. Cells differing from the previously defined canonical nucleolar structure (Ogbadoyi et al., 2000) (Figure 6A) were scored as abnormal. A variety of abnormal nucleoli were observed, the majority of which fell into two categories: 1) unusual heterogeneous density and 2) the appearance of an intranucleolar electron dense ring (Figure 6B). Analysis of the resultant counts showed that abnormal nucleoli were only found in TbRPB5z RNAi cells (Figure 6B).
Figure 6.
Effect of RNAi on structure of the nucleolus. (A) Wild-type homogenous density nucleolus. (B) Examples of nucleoli with heterogenous density and electron-dense ring from TbRPB5z RNAi-induced cells. Numbers below images are percentages from counts of RNAi-induced cells.
DISCUSSION
Nuclear gene transcription in eukaryotes is mediated by three classes of DNA-directed RNA polymerase complex: RNA polymerase I (Pol I) and Pol II and Pol III, each composed of a core set of 12 subunits plus additional class-specific components. Several subunits are known to be specific to a particular polymerase complex (or complexes), but five subunits—RPB5, RPB6, RPB8, RPB10, and RPB12—are thought to be common to all 3 polymerase classes. It has been previously noted that there have been expansions in the repertoire of RPB5 subunits encoded by the genomes of higher plants (Larkin et al., 1999), Giardia spp. (Best et al., 2004), and trypanosomes (Ivens et al., 2005; Kelly et al., 2005; Walgraffe et al., 2005), and that for the latter, there has also been an expansion of the RPB6 family (Kelly et al., 2005; Ivens et al., 2005; Nguyen et al., 2006). However, to date, no functional consequence of these expansions has been described.
Here, we have shown that, in addition to the RPB5 expansions mentioned above, there has also been a duplication of RPB5 in the ciliate P. tetraurelia and that, taken together, these most likely represent four independent expansions of the RPB5 repertoire in the four distinct eukaryotic lineages. We also identify duplications in RPB6 in higher plants, trypanosomes, and P. tetraurelia (but not in G. lamblia). Again, these most likely represent independent expansions in the different lineages and not an ancestral eukaryotic state. These data show that elaboration of the “common” polymerase subunits—and in particular RPB5 and RPB6—is a recurrent theme in eukaryote polymerase evolution and that the presence of paralogues could have implications in transcriptional function in several eukaryotic lines.
We investigated the function of different paralogues of RPB5 and RPB6 by using as a model system the parasite T. brucei. Nuclear transcription in this organism is particularly interesting for two reasons. First, transcription of protein-coding genes is polycistronic; a single transcript is converted into multiple mature mRNAs, each of which is capped at the 5' end, by trans-splicing of a specific leader sequence, and polyadenylated at the 3′ end. Second, although Pol II transcribes the majority of protein-coding genes, some highly expressed proteins of the cell surface are transcribed by Pol I. These transcripts are processed to form mature mRNA in a manner similar to that for Pol II transcripts. Moreover, in at least one life cycle stage, this Pol I-mediated transcription occurs not in the nucleolus, but in a transcription factory in the nucleoplasm called the ESB.
On the basis of domain organization, the trypanosomatid paralogues of both RPB5 and RPB6 can be divided into two groups. One group is identical in domain organization to the canonical eukaryotic subunit—these subunits are named RPB5 and RPB6, respectively. The other group differs in domain organization by gain of novel insertions or loss of conserved regions and—these variant subunits are named RPB5z and RPB6z, respectively. By expression of chimeric proteins, we have demonstrated that the canonical forms of both RPB5 and RPB6 are found exclusively in the nucleoplasm of T. brucei and never in the nucleolus. This implies a role for these canonical forms in Pol II and/or Pol III complexes but not in the nucleolar Pol I complex that transcribes the rRNA genes. Conversely, TbRPB5z and TbRPB6z localize exclusively to the nucleolus, and, in bloodstream-form cells, to an additional focus in the nucleoplasm demonstrated to be the Pol I-containing ESB. Tandem affinity purification of complexes containing either RPB5 or RPB5z showed that these two paralogues do indeed interact with different polymerase complexes: RPB5 with both Pol II and Pol III and RPB5z specifically with Pol I. Furthermore, RNAi-mediated knockdown of either RPB5 or RPB5z resulted in a specific inhibition of transcription by either both Pol II and Pol III, or Pol I, respectively. The duplicated RPB5 and RPB6 subunits are the first documented examples of conventionally shared subunits being specific to different polymerase classes. Moreover, TbRPB5 and TbRPB6 are the first examples of polymerase subunits shared between Pol II and Pol III but not Pol I.
Taken together, these data demonstrate that the elaboration of RPB5 and RPB6 in trypanosomes is linked to a specialization of the paralogues to particular polymerase complexes. This elaboration involves the emergence of a noncanonical variant of both subunits that is specific for Pol I. Interestingly, the specificity of the variant forms for Pol I is true for nucleolar rRNA transcription and also for the Pol I-mediated transcription of protein-coding genes in the nucleoplasm. Thus, there seems to be no form of RPB5 or RPB6 that is specific to the ESB or that is required for coupling of mRNA processing to the Pol I complex in T. brucei. Endogenous Pol I-meditated transcription of protein-coding genes has so far not been detected in organisms other than T. brucei. However, it is worth noting that the same elaborations of RPB5 and RPB6 are found in other Kinetoplastidae (for example, T. cruzi and L. major). Moreover, Pol I promoters have been used for the expression of reporter genes in other trypanosomatids such as T. cruzi (Tyler-Cross et al., 1995; dos Santos and Buck, 2000) and L. major (Gay et al., 1996; Boucher et al., 2002) with no apparent aberrations in mRNA processing.
What significance might the elaboration of polymerase subunits found in trypanosomes and other eukaryotic lineages have on the interactions made by the polymerase complex? First, the variant paralogue of RPB6 in trypanosomes has lost the N-terminal domain characteristic of most eukaryotic RPB6 homologues. In this way, this trypanosomatid RPB6z sequence, and also the sole RPB6 subunit of G. lamblia, are similar in structure to the archaeal RPB6 homologue rpoK. Second, in T. brucei, the variant paralogues of RPB5 differs from the canonical structure by the insertion of charged residues. These insertions are in regions predicted to be on the polymerase surface and would considerably alter the interactions made by this region of the polymerase.
It has been shown that S. cerevisiae RPB6 is a point of contact between the RPB4/RPB7 subcomplex and RNA polymerase II and that this contact is mediated by RPB4 (Tan et al., 2003). Given the position of RPB6 in the Pol II complex (Cramer et al., 2000; Kettenberger et al., 2004), this interaction must be mediated by the charged N-terminal domain, for which the orientation and structure are not known. The absence of the charged N-terminal domain from G. lamblia RPB6 and trypanosome RPB6z may therefore affect their ability to recruit the RPB4/RPB7 heterodimer to the polymerase complex. Because trypanosomatids lack the Pol I and Pol III paralogues of RPB4 (Kelly et al., 2005), it is likely that incorporation of RPB6z into Pol I will impede subsequent recruitment of the RPB4/RPB7 heterodimer, which is known to play a role in many aspects of the transcription cycle (Sampath and Sadhale, 2005). The insertion in TbRPB6z that disrupts the C-terminal domain (which is also found in the apicomplexan parasites P. falciparum and T. annulata) might enable these proteins to interact with Pol I-specific transcription factors.
RPB5 forms part of the DNA entrance channel in the solved structure of yeast Pol II (Cramer et al., 2000). As such, it is in a strategic position for interaction with DNA-bound transcription-associated factors. In support of this, RPB5 has been shown to interact with a number of transcriptional regulators, including TFIIB (Lin et al., 1997), TFIIF (Wei et al., 2001) and TFIIE (Hayashi et al., 2005). Diversification of sequence within this subunit, hence changing the interacting face of the polymerase, is therefore a simple way in which an organism might alter the function of an entire polymerase complex. By homology modeling of T. brucei RPB5z, we predict that the N-terminal insertions form surface-exposed structures. The most N-terminal of these insertions is predicted to form a novel face for the polymerase, directly adjacent to the DNA entrance channel. This novel front would greatly alter its ability to interact with transcription factors that usually make contact through RPB5. It will be interesting to see whether the multiple paralogues of both RPB5 and RPB6 found in other eukaryotes are also specific to different polymerase classes, as we have shown for T. brucei, and what role they will play in the differential regulation of gene expression mediated by these complexes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Jeff Errington (Newcastle University, United Kingdom) for the gift of polyclonal rabbit anti-GFP antibody and also Jean-François Dierick (Biovallée, Charleroi, Belgium) for the mass-spectrometry analysis. Predicted protein datasets were obtained from the sources specified in Supplemental Material. We thank each of these organizations and the respective genome sequencing projects for making sequence, gene model, and annotation data publicly available. S.D. is a recipient of a fellowship from the Belgian Fonds pour la formation à la Recherché dans l'Industrie et dans l'Agriculture. L.V. is Maître de Recherches at the Belgian Fonds national de la Recherche Scientifique. In E.P.'s lab this work was supported by grants from the Belgian Fonds de la Recherche Fondamentale (to E.P.), the Belgian Fonds de la Recherche Scientifique Médicale (to L.V. and E.P.), a Crédit aux Chercheurs from the Belgian Fonds national de la Recherche Scientifique (to L.V.), the Human Frontier Scientific Program (to L.V.), and the Interuniversity Attraction Poles Programme – Belgian Science Policy (to E.P.). In K.G.'s lab this work was supported by the Wellcome Trust, the Human Frontier Science Program, and the EP Abraham Rrust. K.G. is a Wellcome Trust Principal Research Fellow.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0841) on January 31, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O., Cain S., Cohen J., Sperling L. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 2007;35:D439–D444. doi: 10.1093/nar/gkl777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best A. A., Morrison H. G., McArthur A. G., Sogin M. L., Olsen G. J. Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia. Genome Res. 2004;14:1537–1547. doi: 10.1101/gr.2256604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher N., McNicoll F., Dumas C., Papadopoulou B. RNA polymerase I-mediated transcription of a reporter gene integrated into different loci of Leishmania. Mol. Biochem. Parasitol. 2002;119:153–158. doi: 10.1016/s0166-6851(01)00410-8. [DOI] [PubMed] [Google Scholar]
- Brun R., Jenni L. A new semi-defined medium for Trypanosoma brucei sspp. Acta Trop. 1977;34:21–33. [PubMed] [Google Scholar]
- Buhler J. M., Huet J., Davies K. E., Sentenac A., Fromageot P. Immunological studies of yeast nuclear RNA polymerases at the subunit level. J. Biol. Chem. 1980;255:9949–9954. [PubMed] [Google Scholar]
- Carruthers V. B., Cross G. A. High-efficiency clonal growth of bloodstream- and insect-form Trypanosoma brucei on agarose plates. Proc. Natl. Acad. Sci. USA. 1992;89:8818–8821. doi: 10.1073/pnas.89.18.8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C. E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Bushnell D. A., Fu J., Gnatt A. L., Maier-Davis B., Thompson N. E., Burgess R. R., Edwards A. M., David P. R., Kornberg R. D. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- Devaux S., Lecordier L., Uzureau P., Walgraffe D., Dierick J. F., Poelvoorde P., Pays E., Vanhamme L. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;148:60–68. doi: 10.1016/j.molbiopara.2006.02.020. [DOI] [PubMed] [Google Scholar]
- dos Santos W. G., Buck G. A. Simultaneous stable expression of neomycin phosphotransferase and green fluorescence protein genes in Trypanosoma cruzi. J. Parasitol. 2000;86:1281–1288. doi: 10.1645/0022-3395(2000)086[1281:SSEONP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Eddy S. R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Gay L. S., Wilson M. E., Donelson J. E. The promoter for the ribosomal RNA genes of Leishmania chagasi. Mol. Biochem. Parasitol. 1996;77:193–200. doi: 10.1016/0166-6851(96)02594-7. [DOI] [PubMed] [Google Scholar]
- Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L. C., Chung H. M., Lee P. T., Lee M. G. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003;2:542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Watanabe T., Tanaka A., Furumoto T., Sato-Tsuchiya C., Kimura M., Yokoi M., Ishihama A., Hanaoka F., Ohkuma Y. Studies of Schizosaccharomyces pombe TFIIE indicate conformational and functional changes in RNA polymerase II at transcription initiation. Genes Cells. 2005;10:207–224. doi: 10.1111/j.1365-2443.2005.00833.x. [DOI] [PubMed] [Google Scholar]
- Hirumi H., Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ivens A. C., et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S., Wickstead B., Gull K. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem. Soc. Trans. 2005;33:1435–1437. doi: 10.1042/BST0331435. [DOI] [PubMed] [Google Scholar]
- Kettenberger H., Armache K. J., Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Lanzendorfer M., Smid A., Klinger C., Schultz P., Sentenac A., Carles C., Riva M. A shared subunit belongs to the eukaryotic core RNA polymerase. Genes Dev. 1997;11:1037–1047. doi: 10.1101/gad.11.8.1037. [DOI] [PubMed] [Google Scholar]
- Larkin R. M., Hagen G., Guilfoyle T. J. Arabidopsis thaliana RNA polymerase II subunits related to yeast and human RPB5. Gene. 1999;231:41–47. doi: 10.1016/s0378-1119(99)00090-6. [DOI] [PubMed] [Google Scholar]
- Le T. T., Zhang S., Hayashi N., Yasukawa M., Delgermaa L., Murakami S. Mutational analysis of human RNA polymerase II subunit 5 (RPB5): the residues critical for interactions with TFIIF subunit RAP30 and hepatitis B virus X protein. J. Biochem. 2005;138:215–224. doi: 10.1093/jb/mvi119. [DOI] [PubMed] [Google Scholar]
- LeBowitz J. H., Smith H. Q., Rusche L., Beverley S. M. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- Lin Y., Nomura T., Cheong J., Dorjsuren D., Iida K., Murakami S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J. Biol. Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M., et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Navarro M., Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- Nguyen T. N., Schimanski B., Zahn A., Klumpp B., Gunzl A. Purification of an eight subunit RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;149:27–37. doi: 10.1016/j.molbiopara.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Ogbadoyi E., Ersfeld K., Robinson D., Sherwin T., Gull K. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma. 2000;108:501–513. doi: 10.1007/s004120050402. [DOI] [PubMed] [Google Scholar]
- Palenchar J. B., Bellofatto V. Gene transcription in trypanosomes. Mol. Biochem. Parasitol. 2006;146:135–141. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Roeder R. G., Rutter W. J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- Roeder R. G., Schwartz L. B., Sklar V. E. Function, structure, and regulation of eukaryotic nuclear RNA polymerases. Symp. Soc. Dev. Biol. 1976:29–52. [PubMed] [Google Scholar]
- Sampath V., Sadhale P. Rpb4 and Rpb 7, a sub-complex integral to multi-subunit RNA polymerases performs a multitude of functions. IUBMB Life. 2005;57:93–102. doi: 10.1080/15216540500078905. [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M. C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit. Rev. Biochem. 1985;18:31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Shpakovski G. V., Acker J., Wintzerith M., Lacroix J. F., Thuriaux P., Vigneron M. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L. Sunderland, MA: Sinauer; 1998. Phylogenetic Analysis by Parsimony (*and Other Methods) [Google Scholar]
- Tan Q., Prysak M. H., Woychik N. A. Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II, and III. Mol. Cell Biol. 2003;23:3329–3338. doi: 10.1128/MCB.23.9.3329-3338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira S. M., daRocha W. D. Control of gene expression and genetic manipulation in the Trypanosomatidae. Genet. Mol. Res. 2003;2:148–158. [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todone F., Weinzierl R. O., Brick P., Onesti S. Crystal structure of RPB5, a universal eukaryotic RNA polymerase subunit and transcription factor interaction target. Proc. Natl. Acad. Sci. USA. 2000;97:6306–6310. doi: 10.1073/pnas.97.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Cross R. E., Short S. L., Floeter-Winter L. M., Buck G. A. Transient expression mediated by the Trypanosoma cruzi rRNA promoter. Mol. Biochem. Parasitol. 1995;72:23–31. doi: 10.1016/0166-6851(95)00061-5. [DOI] [PubMed] [Google Scholar]
- Ullu E., Matthews K. R., Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell. Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 1990;18:3319–3326. doi: 10.1093/nar/18.11.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L., Pays E. Control of gene expression in trypanosomes. Microbiol. Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walgraffe D., Devaux S., Lecordier L., Dierick J. F., Dieu M., Van den Abbeele J., Pays E., Vanhamme L. Characterization of subunits of the RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 2005;139:249–260. doi: 10.1016/j.molbiopara.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris J. C., Drew M. E., Englund P. T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wei W., Dorjsuren D., Lin Y., Qin W., Nomura T., Hayashi N., Murakami S. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J. Biol. Chem. 2001;276:12266–12273. doi: 10.1074/jbc.M009634200. [DOI] [PubMed] [Google Scholar]
- Wickstead B., Ersfeld K., Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G. A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.