Abstract
Translation of the X-linked inhibitor of apoptosis (XIAP) proceeds by internal ribosome entry site (IRES)-mediated initiation, a process that is physiologically important because XIAP expression is essential for cell survival under conditions of compromised cap-dependent translation, such as cellular stress. The regulation of internal initiation requires the interaction of IRES trans-acting factors (ITAFs) with the IRES element. We used RNA-affinity chromatography to identify XIAP ITAFs and isolated the heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). We find that hnRNP A1 interacts with XIAP IRES RNA both in vitro and in vivo and that hnRNP A1 negatively regulates XIAP IRES activity. Moreover, XIAP IRES-dependent translation is significantly reduced when hnRNP A1 accumulates in the cytoplasm. Osmotic shock, a cellular stress that causes cytoplasmic accumulation of hnRNP A1, also leads to a decrease in XIAP levels that is abrogated by knockdown of hnRNP A1 expression. These results suggest that the subcellular localization of hnRNP A1 is an important determinant of its ability to negatively regulate XIAP IRES activity, suggesting that the subcellular distribution of ITAFs plays a critical role in regulating IRES-dependent translation. Our findings demonstrate that cytoplasmic hnRNP A1 is a negative regulator of XIAP IRES-dependent translation, indicating a novel function for the cytoplasmic form of this protein.
INTRODUCTION
The majority of protein translation in eukaryotic cells is initiated using a 7-methylguanosine cap-dependent ribosome-scanning mechanism (reviewed in Kapp and Lorsch, 2004). However, under conditions of cellular stress, cap-dependent translation is compromised due to the modification or proteolytic degradation of several proteins that are required for translation initiation. For example, endoplasmic reticulum stress causes phosphorylation of eIF2-α, which decreases the rate of cap-dependent translation initiation (Harding et al., 1999). Caspase-mediated cleavage of eIF4G family members, which form a scaffold on which the translation initiation complex assembles, reduces cap-dependent translation during apoptosis (Marissen and Lloyd, 1998). Thus, for translation of many mRNA transcripts to continue under such conditions these messages rely on an alternative initiation mechanism.
Internal ribosome entry sites (IRESs) are believed to permit translation initiation by directly recruiting the ribosome to the vicinity of the start codon, thereby bypassing the need for cap-binding and ribosome scanning (Hellen and Sarnow, 2001). Moreover, IRES elements allow translation of a subset of proteins to persist under conditions of reduced cap-dependent translation, such as apoptosis (Holcik et al., 2000; Hellen and Sarnow, 2001). Translation of the X-linked inhibitor of apoptosis (XIAP), an antiapoptotic protein that binds to caspases-3, -7, and -9 to inhibit their activity (reviewed in Liston et al., 2003), is mediated by a 162 nucleotide IRES element located within its 1.7 kb-long 5′ untranslated region, thereby allowing de novo XIAP synthesis during cellular stress and apoptosis (Holcik et al., 1999). Thus, by exploiting an IRES-dependent translation mechanism, a new XIAP protein can be produced when it is required to block or delay the progression of apoptosis (Holcik and Sonenberg, 2005; Lewis and Holcik, 2005).
Several proteins that modulate IRES function, known as IRES trans-acting factors (ITAFs), have recently been identified (reviewed in Spriggs et al., 2005). These ITAFs interact with various IRES elements to regulate their activity by affecting ribosome recruitment or modifying the structure of the IRES RNA itself. For example, the La autoantigen, PTB, unr, and hnRNP C1/C2 stimulate the activity of several IRES elements (Holcik and Korneluk, 2000; Kim et al., 2001; Mitchell et al., 2001; Holcik et al., 2003), whereas HuR inhibits p27 IRES activity (Kullmann et al., 2002). The XIAP ITAFs identified thus far are the La autoantigen and hnRNP C1/C2 (Holcik and Korneluk, 2000; Holcik et al., 2003); however, the identities of additional protein factors that modulate XIAP IRES function remain unknown.
We have used an RNA-affinity chromatography approach to isolate and identify novel XIAP IRES-binding proteins. We find that the heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) interacts with XIAP IRES RNA both in vitro and in vivo and that hnRNP A1 negatively regulates XIAP IRES activity. Significantly, hnRNP A1–mediated negative regulation of XIAP IRES activity is controlled by the subcellular localization of hnRNP A1, as XIAP IRES-dependent translation is significantly reduced when hnRNP A1 accumulates in the cytoplasm. On the basis of our findings, we conclude that hnRNP A1 is a novel negative regulator of IRES-mediated XIAP translation, whose activity is regulated by subcellular distribution of the protein.
MATERIALS AND METHODS
Cell Culture and Reagents
Human embryonic kidney (HEK293T) cells were cultured in standard conditions in DMEM supplemented with 10% fetal calf serum, glutamate, and antibiotics. Transient transfections were performed using LipofectAMINE PLUS according to the protocol provided by the manufacturer (Invitrogen, Carlsbad, CA). Briefly, cells were seeded at a density of 6 × 105 cells/well in six-well plates and were transfected 24 h later in serum-free DMEM with 2 μg DNA. The transfection mixture was supplemented 3 h later with 1 ml DMEM containing 10% fetal calf serum, glutamate, and antibiotics. Cells were collected for analysis 48 h after transfection. siRNA transfections were performed using RNAifect according to the protocol provided by the manufacturer (Qiagen, Chatsworth, CA). Briefly, cells were seeded at a density of 2.5 × 105 cells/well in six-well plates and were transfected 24 h later in serum-free DMEM with a 20 nM final concentration of hnRNP A1 siRNA (Santa Cruz Biotechnology, Santa Cruz, CA) or a nonsilencing control siRNA (Qiagen). Cells were collected for analysis 72 h after transfection.
The bicistronic reporter plasmid pβgal/5′(−162)/CAT containing the minimal functional region of the human XIAP IRES was previously described (Holcik et al., 1999). The hnRNP A1 coding sequence was PCR amplified from cDNA generated by reverse transcription of total RNA from 293T cells using oligo dT18 primer and the Bulk 1st-Strand Synthesis kit (Amersham Biosciences, Piscataway. NJ). The FLAG epitope was incorporated into the N-terminus of hnRNP A1 using the following primers: 5′-ccggaattcatggattacaaggacgacgacgataagtctaagtcagagtctcctaaagag (EcoRI recognition site is underlined; FLAG coding sequence is italicized) and 5′-tgctctagattaaaatcttctgccactgcc (XbaI recognition site is underlined). The resulting PCR product was cloned into the pCI vector (Invitrogen) using EcoRI and XbaI to generate the FLAG-hnRNP A1 plasmid. A plasmid for expressing recombinant glutathione S-transferase (GST)-hnRNP A1 was generated by PCR amplification of the hnRNP A1 coding sequence (without the AUG start codon) using the primers 5′-ccggaattctatctaagtcagagtctcctaaagag (EcoRI recognition site is underlined) and 5′-ccgctcgagttaaaatcttctgccactgcc (XhoI recognition site is underlined). The resulting PCR product was digested with EcoRI and XhoI and cloned into the pGEX-KG vector. A plasmid expressing FLAG-tagged hnRNP A1 F1 mutant was generated by mutation of the F-peptide within hnRNP A1 as previously described (Allemand et al., 2005). Briefly, hnRNP A1 F1 mutant sequence was amplified with the primers: 5′-ccggaattcatggattacaaggacgacgacgataagtctaagtcagagtctcctaaagag (EcoRI recognition site is underlined; FLAG coding sequence is italicized) and 5′-tgctctagattaaaatcttctgccgtcgccataatcgtcatcgtcatcgtcaccgccatagccaccttggtttcgtgg (XbaI recognition site is underlined; base pair changes to generate the F1 mutant are italicized). The resulting PCR product was cloned into the pCI vector (Invitrogen) using EcoRI and XbaI. All plasmid constructs were confirmed by nucleotide sequencing.
RNA-Affinity Chromatography
Isolation of XIAP IRES-binding proteins was performed using a modified RNA-affinity chromatography protocol (Kim et al., 2004). Briefly, XIAP IRES RNA and HIAP2 IRES RNA were transcribed in vitro with the MEGAShortscript transcription kit according to the manufacturer's protocol (Ambion, Austin, TX) and were biotinylated at the 5′ end with the 5′ EndTag Nucleic Acid Labeling System according to the manufacturer's instructions (Vector Laboratories, Burlingame, CA). The biotinylated RNAs (60 μg) were conjugated to Avidin-agarose beads (Sigma, St. Louis, MO) in the presence of incubation buffer (10 mM Tris-Cl, pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride, 0.05% [vol/vol] Nonidet P-40) at 4°C for 2 h with continuous rotation. Unbound RNAs were removed by washing beads twice with incubation buffer. 293T protein extract (2 mg in incubation buffer) was added to the coated beads, along with 120 μg yeast tRNA (Sigma) and 800 U of Prime RNase inhibitor (Eppendorf, Fremont, CA). Reactions were incubated at room temperature with continuous rotation for 30 min, followed by incubation at 4°C with continuous rotation for 2 h. Beads were washed five times with incubation buffer, resuspended in 50 μl of 1× SDS-PAGE loading dye, and boiled for 5 min to elute bound proteins. Proteins were separated by 10% SDS-PAGE and visualized using Sypro Ruby stain (Genomic Solutions, Ann Arbor, MI). Protein bands were excised and identified by in-gel trypsin digestion and mass peptide fingerprinting at the Protein Function Discovery Centre (Queen's University, Kingston, ON, Canada).
RNA–Protein Complex Immunoprecipitation
In vivo cross-linking and coprecipitation of RNA–protein complexes was performed as described previously (Niranjanakumari et al., 2002). Cross-linked RNA–protein complexes were immunoprecipitated using anti-hnRNP A1 (Santa Cruz Biotechnology), anti-La (Chan and Tan, 1987), or anti-GAPDH (Advanced ImmunoChemical, Long Beach, CA) antibodies at 1:50 dilution. After immunoprecipitation and cross-link reversal, RNA was isolated using Trizol reagent following the manufacturer's protocol (Invitrogen). cDNA was generated from the isolated RNA using an oligo dT18 primer and Superscript II (Invitrogen). The partial coding sequences of XIAP and actin were PCR amplified from the resulting cDNA using the primers 5′-gcggtgctttagttgtcat (XIAP forward), 5′-tcgggtatatggtgtctgata (XIAP reverse), 5′-ctggaacggtgaaggtgaca (actin forward), and 5′-aagggacttcctgtaacaatgca (actin reverse). PCR products were visualized on a 1.5% agarose gel by ethidium bromide staining.
UV Cross-Linking of RNA–Protein Complexes
RNA–protein UV cross-linking experiments and oligonucleotide competition experiments were performed as previously described (Holcik et al., 2003). For competition experiments, unlabeled XIAP IRES RNA, unlabeled HIAP2 IRES RNA, or unlabeled RNA oligonucleotides were incubated with GST-hnRNP A1 for 15 min before the addition of 32P-labeled XIAP IRES RNA. Sequences of RNA oligonucleotides used for competition experiments are as follows: −25 (5′-ggacaaguccuauuuucaagagaag), −50 (5′-auaauguucucuuuuuagaaaaggu), −75 (5′-uguuucacauuuuggauuuccuaau), and −100 (5′-uccuauaacaaaagucuguugcuug).
Nitrocellulose Filter Bindings Assays
The filter binding assays were performed essentially as described (Bonnal et al., 2005). Briefly, increasing amounts of a purified GST-hnRNP A1 were added to an in vitro transcribed 32P-labeled RNA corresponding to the XIAP IRES or HIAP2 IRES in a total volume of 10 μl GS binding buffer (5 mM HEPES-KOH, pH 7.6, 30 mM KCl, 2 mM MgCl2, 0.2 mM DTT, and 4% glycerol) containing 400 ng of yeast tRNA. The mixture was allowed to incubate 10 min at room temperature. Eight microliters of each binding reaction was applied to a presoaked nitrocellulose membrane on a slot dot apparatus (Hybrislot manifold, Bethesda Research Laboratories, Bethesda, MD) under moderate suction. Each slot dot was washed with 300 μl of cold GS buffer and the membranes were dried for 1 h at room temperature. The filters were exposed in a phosphoimager cassette (Molecular Dynamics, Sunnyvale, CA) overnight and revealed. The quantifications were performed with the Image Quant v1.1 software (Molecular Dynamics), and the data were corrected for the background (RNA retention without any added protein), which was <2%. The fraction of RNA bound was plotted against the protein concentration.
β-Galactosidase and CAT Analysis
Transiently transfected cells were washed in 1 ml phosphate-buffered saline (PBS) and harvested in 300 μl CAT ELISA kit lysis buffer according to the protocol provided by the manufacturer (Roche Molecular Biochemicals, Indianapolis, IN). β-Galactosidase (βgal) enzymatic activity was determined by spectrophotometric assay using o-nitrophenyl-β-d-galactopyranoside as previously described (MacGregor et al., 1991). CAT levels were determined using the CAT ELISA kit according to the protocol provided by the manufacturer (Roche Molecular Biochemicals). Relative IRES activity was calculated as the ratio of CAT/βgal.
Western Blot Analysis
Cells were washed in 1 ml PBS and lysed in 100 μl RIPA buffer for 30 min at 4°C, followed by centrifugation at 12,000 × g for 10 min to pellet debris. Protein concentration was assayed by Bradford (Bio-Rad, Richmond, CA), and equal amounts of protein extract were separated by 10% SDS-PAGE and transferred to PVDF membrane. Samples were analyzed by Western blotting using mouse monoclonal anti-hnRNP A1 (Santa Cruz Biotechnology), rabbit polyclonal anti-XIAP (Aegera Therapeutics, Montreal, Quebec, Canada), or mouse-monoclonal anti-GAPDH (Advanced ImmunoChemical) followed by secondary antibody (horseradish peroxidase–conjugated anti-mouse or anti-rabbit IgG; Amersham Biosciences). Antibody complexes were detected using the ECL or ECL Plus systems (Amersham Biosciences).
Quantitative RT-PCR Analysis
Total RNA was isolated from transfected cells using the Absolutely RNA miniprep kit according to the manufacturer's instructions (Stratagene, La Jolla, CA). cDNA was generated using an oligo dT18 primer and the Bulk 1st-Strand Synthesis kit according to the protocol provided by the manufacturer (Amersham Biosciences). The synthesized cDNA was used as the template for quantitative PCR using the QuantiTect SYBR Green PCR kit (Qiagen) and analyzed on an ABI Prism 7000 detection system (Columbia, MD) using the ABI Prism 7000 SDS software. Quantitative PCR reactions were carried out to detect βgal (forward: 5′-actatcccgaccgccttact; reverse: 5′-ctgtagcgctgatgttgaa), CAT (forward: 5′-gcgtgttacggtgaaaacct; reverse: 5′-gggcgaagaacttgtccata), XIAP (forward: 5′-gcggtgctttagttgtcat; reverse: 5′-tcgggtatatggtgtctgata), or GAPDH (forward: 5′-acagtcagccgcatcttctt; reverse: 5′-acgaccaaatccgttgactc).
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described (Allemand et al., 2005). FLAG-tagged proteins were detected using mouse monoclonal anti-FLAG M2 (Sigma) and Alexa Fluor 488–conjugated goat anti-mouse IgG secondary antibody (Molecular Probes, Eugene, OR).
XIAP Protein Stability Analysis
XIAP protein stability was determined essentially as described (Yoon et al., 2006). Cells were treated with 10 μg/ml cycloheximide (Sigma) 18 h after transfection. After 45 min of cycloheximide treatment, protein extracts were harvested by lysis in 100 μl RIPA buffer (0-h time point). Protein extracts were subsequently harvested at 1-, 2-, 4-, and 8-h time points, protein concentration was assayed by Bradford (Bio-Rad), and equal amounts of protein extract were separated by 10% SDS-PAGE, transferred to PVDF membrane, and analyzed by Western blot using antibodies against XIAP and GAPDH.
Subcellular Fractionation
Cells were washed in 1 ml of ice-cold PBS, resuspended in 400 μl of buffer A (10 mM HEPES-KOH, pH 7.5, 10 mM KCl, 1 mM DTT, 1 mM PMSF) containing protease inhibitors, and incubated on ice for 15 min. Nonidet P-40 was added to a final concentration of 0.3% and cells were incubated for an additional 10 min on ice. Nuclei were pelleted by centrifugation at 1500 × g for 5 min, and the cytoplasmic fraction was collected to a new tube and clarified by centrifugation at 13,000 × g for 15 min. The nuclei pellet was washed two times with 1 ml of buffer A, then resuspended in 50 μl of buffer B (20 mM HEPES-KOH, pH 7.5, 400 mM NaCl, 1 mM DTT, 1 mM PMSF) containing protease inhibitors, and incubated on ice for 30 min, with mixing every 5 min. Nuclear debris was pelleted at 13,000 × g for 5 min, and the nuclear fraction was collected to a new tube.
RESULTS
hnRNP A1 Interacts with the XIAP IRES
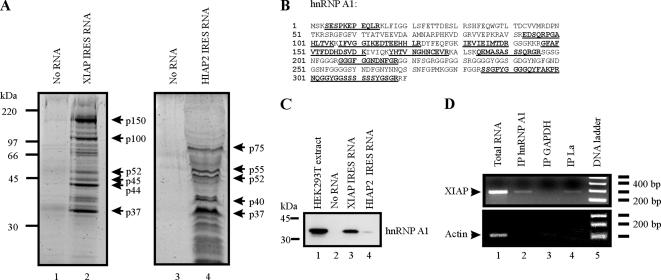
We previously demonstrated that the La autoantigen and hnRNP C1/C2 interact with the XIAP IRES and modulate its ability to support translation (Holcik and Korneluk, 2000; Holcik et al., 2003). To gain a better understanding of XIAP IRES function, we used an RNA-affinity chromatography approach (Kim et al., 2004) followed by mass-peptide fingerprinting to isolate and identify proteins that interact with XIAP IRES RNA. We used the minimal functional XIAP IRES element that retains full activity and encompasses nucleotides −162 to +1 in the 5′ untranslated region of XIAP mRNA for these experiments (Holcik et al., 1999). Whole-cell protein extract was prepared from HEK293T cells, which was incubated with either avidin-agarose beads alone, avidin-agarose beads conjugated to biotin-labeled XIAP IRES RNA, or avidin-agarose beads conjugated to biotin-labeled HIAP2 IRES RNA (the 149-nucleotide minimal functional IRES sequence; Warnakulasuriyarachchi et al., 2004). When XIAP IRES RNA was used as an affinity matrix, we were able to isolate at least six distinct proteins (Figure 1A, lane 2), whereas a control reaction using avidin-agarose beads alone did not yield any proteins (Figure 1A, lanes 1 and 3). The use of HIAP2 IRES RNA as an affinity matrix resulted in the isolation of a different set of proteins than that isolated using XIAP IRES RNA (Figure 1A, compare lane 4 to lane 2), indicating that the proteins isolated with XIAP IRES RNA were not simply nonspecific RNA-binding proteins. We first subjected the p52 protein isolated with XIAP IRES RNA to mass-peptide fingerprinting because it is the same molecular weight as the La autoantigen, which was previously shown to interact with XIAP IRES and which we therefore expected to isolate using this technique. Indeed, the p52 protein was identified as the La autoantigen, confirming that we were able to isolate XIAP IRES-binding proteins using an RNA-affinity chromatography approach (data not shown). Similarly, we also isolated hnRNP C1/C2 using this technique, as determined by Western blot analysis (p44 protein; data not shown). We then subjected the p37 protein isolated using XIAP IRES RNA to mass-peptide fingerprinting and identified it as hnRNP A1 with 42% sequence coverage (Figure 1B).
Figure 1.
hnRNP A1 interacts with the XIAP IRES. (A) RNA-affinity chromatography isolation of XIAP IRES-binding proteins. Precleared protein extracts from HEK293T cells were incubated with either agarose beads coated with XIAP IRES RNA, agarose beads coated with HIAP2 IRES RNA, or agarose beads alone. After protein binding, beads were washed extensively and pelleted, and proteins were eluted by boiling and resolved by SDS-PAGE. Proteins were visualized with Sypro Ruby stain, and the indicated protein species were excised from the gel and subjected to mass spectrometry analysis. (B) The p37 protein is hnRNP A1. Underlined peptide sequences were identified by mass spectrometry analysis of the p37 protein species isolated by RNA-affinity chromatography using XIAP IRES RNA as an affinity matrix. (C) hnRNP A1 associates with XIAP IRES RNA in vitro. XIAP IRES RNA and HIAP2 IRES RNA were used in RNA-affinity chromatography as described in A; isolated proteins were separated by SDS-PAGE, transferred to PVDF membrane, and probed with anti-hnRNP A1 antibody. (D) hnRNP A1 associates with XIAP mRNA in vivo. RNA–protein complexes were cross-linked with formaldehyde, isolated from cells, and immunoprecipitated using antibodies against hnRNP A1, La, and GAPDH. After immunoprecipitation, RNA–protein cross-links were reversed, and the RNA was isolated and used in an RT-PCR reaction with XIAP- and actin-specific oligonucleotide primers.
To confirm that hnRNP A1 associates with the XIAP IRES, we performed RNA-affinity chromatography and subjected the isolated protein sample to Western blot analysis using hnRNP A1 antibodies. Again, we included HIAP2 IRES RNA as an affinity matrix to test the specificity of the hnRNP A1 interaction with the XIAP IRES. hnRNP A1 was detected in the protein eluate from XIAP IRES RNA affinity matrix, but was not detected in the protein eluate from avidin-agarose beads alone (Figure 1C, compare lane 3 to lane 2). Moreover, hnRNP A1 was only very weakly associated with HIAP2 IRES RNA (Figure 1C, lane 4), demonstrating that hnRNP A1 does not interact nonspecifically with all IRES (or RNA) sequences. Therefore, hnRNP A1 specifically associates with XIAP IRES RNA in vitro.
We next assessed whether the endogenous hnRNP A1 associates with endogenous XIAP mRNA in cells. RNA–protein complexes were formaldehyde cross-linked in whole cells, and after cell lysis RNA–protein complexes were immunoprecipitated, and RNA was isolated from these immunoprecipitates. cDNA was produced by reverse transcription, followed by PCR amplification with XIAP and actin-coding sequence-specific primers. As shown previously (Holcik and Korneluk, 2000), we were able to isolate XIAP mRNA by immunoprecipitation with La antibodies (Figure 1D, lane 4) but not with GAPDH antibodies (negative control, Figure 1D, lane 3). Immunoprecipitation with hnRNP A1 antibodies coprecipitated XIAP mRNA (Figure 1D, lane 2), confirming that endogenous hnRNP A1 associates with endogenous XIAP mRNA in cells in vivo. Importantly, we were unable to amplify the high-abundance actin transcript from our immunoprecipitates, indicating that our coimmunoprecipitation of XIAP mRNA is specific.
hnRNP A1 Binds within the Core RNP-Binding Site of the XIAP IRES
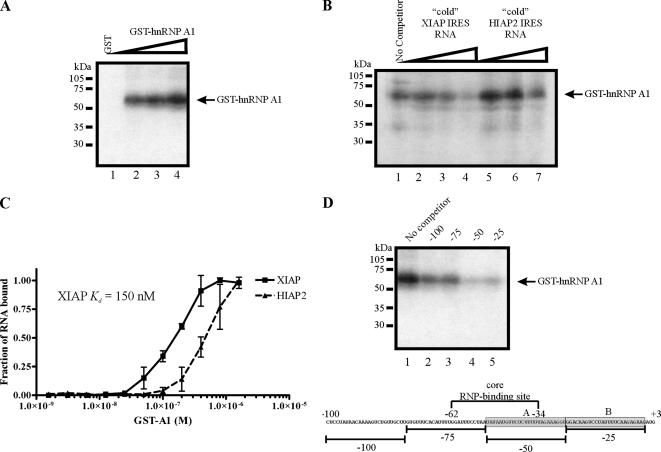
Our observations that hnRNP A1 is associated with XIAP IRES RNA both in vitro and in vivo suggest that hnRNP A1 can bind directly to the XIAP IRES sequence. To determine if hnRNP A1 binds directly to XIAP IRES RNA, we performed a UV-cross-linking experiment using a radiolabeled XIAP IRES RNA probe and purified recombinant GST-hnRNP A1. Increasing amounts of recombinant GST-hnRNP A1 were incubated with 32P-labeled XIAP IRES RNA, followed by UV cross-linking and separation by SDS-PAGE. We find that XIAP IRES RNA is cross-linked to GST-hnRNP A1 in vitro (Figure 2A), indicating that hnRNP A1 does indeed bind directly to XIAP IRES RNA. To test the specificity of this interaction, we coincubated GST-hnRNP A1 and 32P-labeled XIAP IRES RNA with excess unlabeled XIAP IRES RNA (specific competitor) or excess unlabeled HIAP2 IRES RNA (nonspecific competitor) in our UV cross-linking assay. We find that whereas excess unlabeled XIAP IRES RNA effectively competes with 32P-labeled XIAP IRES RNA for binding to GST-hnRNP A1 (Figure 2B, lanes 3 and 4), excess unlabeled HIAP2 IRES RNA does not (Figure 2B, lanes 6 and 7). Therefore, the in vitro binding of GST-hnRNP A1 to XIAP IRES RNA is specific.
Figure 2.
hnRNP A1 binds directly to XIAP IRES RNA within the core RNP-binding site. (A) GST (lane 1) or increasing amounts of GST-hnRNP A1 (lanes 2–4) were incubated with 32P-labeled XIAP IRES RNA probe, UV-cross-linked, and then separated by SDS-PAGE and visualized by autoradiography. (B) GST-hnRNPA1 was incubated with 32P-labeled XIAP IRES RNA probe alone (No competitor) or a combination of 32P-labeled XIAP IRES RNA probe and increasing amounts of excess unlabeled (“cold”) competitor RNA molecules (XIAP IRES RNA and HIAP2 IRES RNA), UV-cross-linked, separated by SDS-PAGE, and visualized by autoradiography. (C) hnRNP A1 binding curve for XIAP IRES and HIAP2 IRES RNA. Nitrocellulose filter binding assays were performed and analyzed as described in Materials and Methods. Filter-bound RNA (■, XIAP IRES; ▴, HIAP2 IRES) is plotted as a function of protein concentration. The data presented represent the mean ± SD of three independent experiments. (D) GST-hnRNPA1 was incubated with 32P-labeled XIAP IRES RNA probe alone (No competitor) or a combination of 32P-labeled XIAP IRES RNA probe and 100-fold excess of unlabeled competitor RNA molecules, UV-cross-linked, separated by SDS-PAGE, and visualized by autoradiography. Gray boxes in the sequence schematic indicate hnRNP A1–binding sites (labeled as site A and site B).
We further assessed the interaction between hnRNP A1 and the XIAP IRES by measuring the apparent equilibrium dissociation constant (Kd) of the recombinant GST-hnRNP A1 protein and XIAP IRES RNA. To determine the Kd of this interaction we performed a nitrocellulose filter binding assay, in which varying amounts of GST-hnRNP A1 are incubated with a constant amount of RNA. We also included HIAP2 IRES RNA as a control, as we have found that hnRNP A1 does not effectively bind to this RNA (Figures 1C and 2B). As shown in Figure 2C, we find the Kd for the interaction between GST-hnRNP A1 and XIAP IRES RNA is 150 nM, whereas the Kd for an interaction between GST-hnRNP A1 and HIAP2 IRES RNA is greater than 480 nM (we were unable to determine the actual value because we were unable to saturate binding). Importantly, the Kd value for the interaction between hnRNP A1 and XIAP IRES RNA is similar to the Kd value previously reported for the interaction between hnRNP A1 and the fibroblast growth factor (FGF)-2 IRES (200 nM; Bonnal et al., 2005), suggesting a bona fide and specific interaction.
The core RNP-binding site within the XIAP IRES is located between nucleotides −62 to −34 (Holcik and Korneluk, 2000). Therefore, we mapped the binding of hnRNP A1 using competitor oligonucleotides as described previously (Holcik and Korneluk, 2000). RNA oligonucleotide competitors that span the polypyrimidine tract within the core RNP-binding site and sequences immediately upstream of the start codon compete with 32P-labeled XIAP IRES RNA for binding to GST-hnRNP A1 (Figure 2D, lanes 4 and 5), indicating that hnRNP A1 binds these regions of the IRES sequence. These observations suggest that hnRNP A1 may modulate XIAP IRES activity by binding in the proximity of the core RNP-binding site, as has been previously observed for the La autoantigen (Holcik and Korneluk, 2000). We additionally tested whether mutations of the identified binding sites within the XIAP IRES abrogate the effect of hnRNP A1 on XIAP IRES activity (see below).
Cytoplasmic hnRNP A1 Negatively Regulates IRES-dependent Translation of XIAP
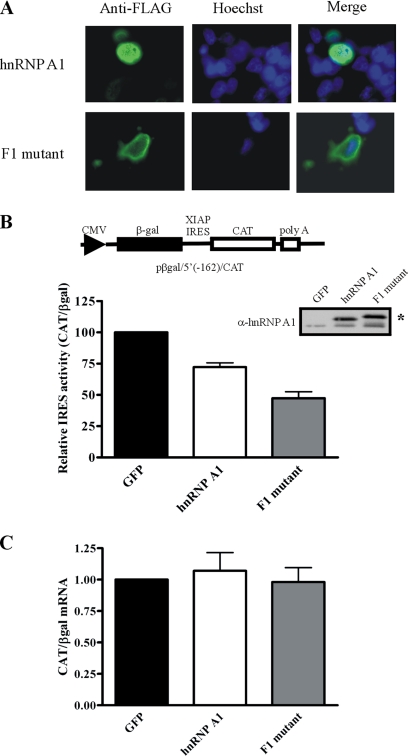
We have shown that hnRNP A1 can interact with the XIAP IRES both in vitro and in vivo, suggesting that hnRNP A1 may modulate XIAP IRES activity. Therefore, we examined whether overexpression of hnRNP A1 has any effect on XIAP IRES activity. To assess XIAP IRES activity, we used a previously described bicistronic reporter plasmid containing the minimal XIAP IRES element (pβgal/5′(−162)/CAT; Holcik et al., 1999); translation of the first cistron (βgal) is cap-dependent whereas translation of the second cistron (CAT) is dependent on XIAP IRES activity (Holcik et al., 1999). By calculating the ratio of CAT expression to βgal expression, the relative IRES activity can be determined. HEK293T cells were transiently cotransfected with the pβgal/5′(−162)/CAT bicistronic reporter plasmid and either a FLAG-tagged hnRNP A1 or green fluorescent protein (GFP)-expressing plasmid. We found that overexpression of FLAG-hnRNP A1 causes a decrease in XIAP IRES activity compared with a GFP control (Figure 3B), indicating that hnRNP A1 has a negative effect on XIAP IRES function. We next determined the effect of mutating the hnRNP A1 binding sites within the XIAP IRES using the same bicistrionic reporter approach. hnRNP A1 binds within the polypyrimidine tract contained in the core RNP-binding site of the XIAP IRES (Figure 2); we have found that the introduction of mutations within this sequence completely abrogates XIAP IRES activity (data not shown and Holcik et al., 1999), precluding us from assessing the effect of mutating this site on hnRNP A1 ITAF function. However, removal of the hnRNP A1 binding site adjacent to the start codon did not affect hnRNP A1 ITAF function (data not shown), suggesting that the hnRNP A1 binding site within the polypyrimidine tract may be sufficient for the negative regulation of XIAP IRES activity by hnRNP A1 in vivo.
Figure 3.
Cytoplasmic localization of hnRNP A1 reduces XIAP IRES activity. (A) Immunohistochemistry analysis of HEK293T cells transfected with FLAG-hnRNP A1 or the FLAG-hnRNP A1 F1 mutant using anti-FLAG antibodies; nuclei were visualized by Hoechst staining. (B) hnRNP A1 reduces XIAP IRES activity. HEK293T cells were cotransfected with a plasmid expressing GFP, FLAG-hnRNP A1, or FLAG-hnRNP A1 F1 mutant and the pβgal/5′(−162)/CAT bicistronic reporter plasmid. 48 h after transfection βgal and CAT protein expression was assayed; relative IRES activity is expressed as a ratio of CAT/βgal. The activity of XIAP IRES in GFP transfected cells was set as 100. Mean ± SEM (bars) of three independent experiments performed in triplicate. Expression levels of FLAG-hnRNP A1 and the FLAG-hnRNP A1 F1 mutant were determined by Western-blot analysis using anti-hnRNP A1 antibodies. The asterisk (*) indicates the FLAG-tagged protein species. (C) Overexpression of hnRNP A1 does not affect the integrity of the bicistronic RNA transcript produced from pβgal/5′(−162)/CAT. Cells were treated as described in B; total RNA was isolated 48 h after transfection, and cDNA was generated by reverse transcription. Quantitative PCR was used to determine the levels of βgal and CAT cistrons; values are expressed as CAT relative to βgal (2−[Ct(CAT) − Ct(βgal)]), and the ratio for GFP-transfected cells was set as 1. Bars, mean ± SD of three independent experiments.
hnRNP A1 has been shown to be involved in alternative splice-site selection (Mayeda and Krainer, 1992), raising the possibility that overexpression of hnRNP A1 affects the integrity of the bicistronic RNA transcript (βgal/IRES/CAT) produced from the pβgal/5′(−162)/CAT plasmid, thus altering the ratio of CAT protein to βgal protein. Therefore we determined the effect of overexpressing hnRNP A1 on the integrity of the βgal/5′(−162)/CAT bicistronic RNA transcript using quantitative RT-PCR as described previously (Holcik et al., 2005). Total RNA was isolated from cells cotransfected with pβgal/5′(−162)/CAT and a GFP- or hnRNP A1–expressing plasmid. cDNA was produced by reverse transcription, which served as a template for quantitative PCR using primers that amplify a portion of the βgal coding region and a portion of the CAT coding region. As shown in Figure 3C, the ratio of CAT and βgal cistrons was unchanged in cells cotransfected with the hnRNP A1–expressing plasmid compared with cells cotransfected with the GFP-expressing plasmid. These data confirm that the integrity of the bicistronic RNA transcript produced from the pβgal/5′(−162)/CAT reporter plasmid is not affected when hnRNP A1 is overexpressed.
hnRNP A1 normally shuttles between the nucleus and the cytoplasm, with the bulk of the protein displaying nuclear localization (Pinol-Roma and Dreyfuss, 1992). We found that overexpressed FLAG-hnRNP A1 is mostly nuclear, although we saw some cytoplasmic accumulation of the protein (Figure 3A). Because translation is a cytoplasmic event, the modest effect of hnRNP A1 overexpression on XIAP IRES-dependent translation may be due to the different subcellular compartmentalization of hnRNP A1 and mRNAs containing the XIAP IRES. Notably, two reports have shown that hnRNP A1 accumulates in the cytoplasm during cellular stress (van der Houven van Oordt et al., 2000; Allemand et al., 2005), suggesting that hnRNP A1 ITAF activity for the XIAP IRES could be regulated by subcellular localization. We therefore hypothesized that hnRNP A1 should exert an inhibitory effect on IRES-dependent translation of XIAP only when hnRNP A1 is present in the cytoplasm. To test this hypothesis, we used a mutant version of hnRNP A1 (the F1 mutant) that fails to interact with the Trn1 transporter protein and thus remains cytoplasmically localized (Figure 3A; Allemand et al., 2005). HEK293T cells were transiently cotransfected with the pβgal/5′(−162)/CAT bicistronic reporter plasmid and a plasmid expressing a FLAG-tagged hnRNP A1 F1 mutant. Although wild-type hnRNP A1 overexpression caused a ∼28% decrease in XIAP IRES activity, the overexpression of the F1 mutant caused a ∼53% decrease in XIAP IRES activity (Figure 3B). Quantitative RT-PCR showed that overexpression of the F1 mutant has no effect on the integrity of the bicistronic reporter RNA (Figure 3C). These data indicate that cytoplasmic hnRNP A1 preferentially affects the activity of the XIAP IRES.
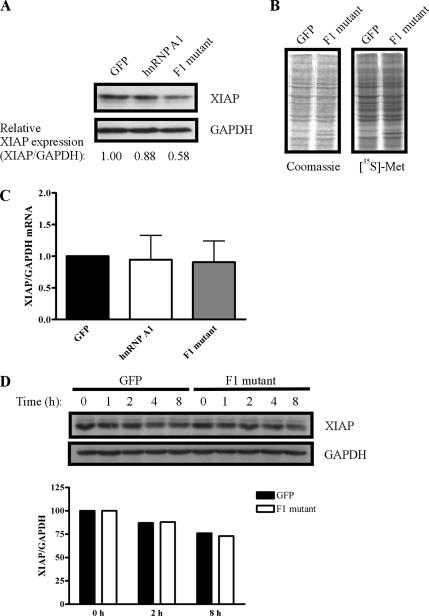
We also examined the effect of cytoplasmic hnRNP A1 on the expression of endogenous XIAP protein. Western blot analysis using anti-XIAP antibodies showed that overexpression of the F1 mutant reduces endogenous XIAP protein levels by nearly half (Figure 4A) but does not affect XIAP mRNA levels (Figure 4C). Importantly, overexpression of the F1 mutant does not affect global protein synthesis (Figure 4B) or the stability of XIAP protein (Figure 4D), indicating that the cytoplasmic hnRNP A1 F1 mutant specifically reduces translation of endogenous XIAP mRNA. Together with the data presented above, these findings demonstrate that cytoplasmic localization of hnRNP A1 is an important determinant of its ability to negatively regulate IRES-dependent translation of XIAP. More importantly, these data provide evidence that subcellular localization of an ITAF affects its ability to modulate IRES function.
Figure 4.
Cytoplasmic localization of hnRNP A1 reduces translation of endogenous XIAP mRNA. (A) Overexpression of hnRNP A1 reduces endogenous XIAP levels. HEK293T cells were transfected with a plasmid expressing GFP, FLAG-hnRNP A1, or FLAG-hnRNP A1 F1 mutant. Forty-eight hours after transfection protein extracts were prepared, separated by SDS-PAGE, transferred to PVDF membrane, and subjected to Western blot analysis with antibodies against XIAP and GAPDH. Relative XIAP levels are expressed as a ratio of XIAP to GAPDH. Displayed data are representative of three independent experiments. (B) Overexpression of the cytoplasmic hnRNP A1 F1 mutant does not affect total protein synthesis. HEK293T cells were transfected with a plasmid expressing GFP or the FLAG-hnRNP A1 F1 mutant, and 24 h later cells were metabolically labeled in the presence of [35S]methionine for 20 min. Protein extracts were harvested, and equal amounts were separated by SDS-PAGE. Proteins were visualized by Coomassie Brilliant Blue staining and autoradiography. (C) Overexpression of hnRNP A1 does not affect XIAP mRNA levels. Cells were treated as described in A; total RNA was isolated 48 h after transfection, and cDNA was generated by reverse transcription. Quantitative PCR was used to determine the levels of XIAP and GAPDH mRNAs; values are expressed as XIAP relative to GAPDH (2−[Ct(XIAP) − Ct(GAPDH)]), and the ratio for GFP-transfected cells was set as 1. Bars, mean ± SD of three experiments. (D) Overexpression of the cytoplasmic hnRNP A1 F1 mutant does not affect XIAP protein stability. Cells were transfected with plasmids expressing GFP or the FLAG-hnRNP A1 F1 mutant. Eighteen hours after transfection cells were treated with 10 μg/ml cyloheximide for 45 min, and then samples were harvested at 0-, 1-, 2-, 4-, and 8-h time points. Equivalent amounts of protein extract were separated by SDS-PAGE, transferred to PVDF, and analyzed by Western blot using antibodies against XIAP and GAPDH. The graph shows XIAP protein levels relative to GAPDH levels (bars), with the 0-h time point set as 100.
Stress-induced Cytoplasmic Accumulation of hnRNP A1 Reduces XIAP Protein Levels
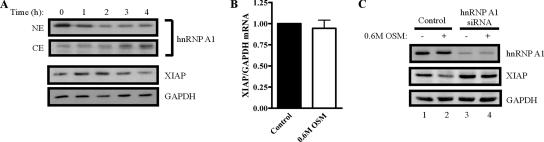
Previous reports have shown that hnRNP A1 accumulates in the cytoplasm of cells exposed to high-osmolarity growth medium (van der Houven van Oordt et al., 2000; Allemand et al., 2005). Because we have observed that cytoplasmic hnRNP A1 significantly reduces XIAP IRES-dependent translation, we hypothesized that osmotic shock should lead to a decrease in XIAP protein levels because of accumulation of hnRNP A1 in the cytoplasm. Moreover, any decrease in XIAP levels caused by osmotic shock should be dependent on hnRNP A1. To determine if osmotic shock reduces XIAP protein levels, cells were grown in media containing 0.6 M sorbitol (osmotic shock medium [OSM]) or nonsupplemented growth media for 4 h, with samples harvested at 1-h intervals. Western blot analysis using anti-XIAP antibodies showed that XIAP protein levels decrease during osmotic shock (Figure 5A). Moreover, this decrease in XIAP protein levels correlates with the redistribution of hnRNP A1 from the nucleus to the cytoplasm during osmotic shock (Figure 5A). Examination of XIAP mRNA levels by quantitative RT-PCR showed that they were not affected by osmotic shock (Figure 5B). Interestingly, we also observed that osmotic shock–mediated cytoplasmic accumulation of hnRNP A1 causes an increase in FGF-2 protein levels (Supplementary Figure S1). FGF-2 is translated using an IRES-dependent mechanism (Vagner et al., 1995), and FGF-2 IRES activity was recently shown to be enhanced by hnRNP A1 (Bonnal et al., 2005). Therefore, our findings suggest that cytoplasmic accumulation of hnRNP A1 is also important for its positive regulation of IRES activity (such as FGF-2 IRES).
Figure 5.
Stress-induced cytoplasmic accumulation of hnRNP A1 reduces XIAP protein levels. (A) Osmotic shock redistributes hnRNP A1 to the cytoplasm and reduces XIAP protein levels. HEK293T cells were grown in high-osmolarity growth medium (0.6 M OSM) or DMEM for 4 h, and nuclear (NE) and cytoplasmic (CE) protein fractions were harvested at 1-h intervals, separated by SDS-PAGE, transferred to PVDF, and subjected to Western blot analysis with antibodies against XIAP, hnRNP A1, and GAPDH. (B) Osmotic shock does not affect XIAP mRNA levels. HEK293T cells were treated with DMEM (Control) or 0.6 M OSM for 4 h, total RNA was then isolated and cDNA was generated by reverse transcription. Quantitative PCR was used to determine the levels of XIAP and GAPDH mRNAs; values are expressed as XIAP relative to GAPDH (2−[Ct(XIAP) − Ct(GAPDH)]), and the ratio for control cells was set as 1. Bars, mean ± SD of three experiments. (C) Reduced XIAP expression during osmotic shock is dependent on hnRNP A1. HEK293T cells were transfected with 20 nM hnRNP A1 siRNA or 20 nM nonsilencing siRNA (control); 72 h later the media was replaced with 0.6 M OSM or DMEM and cells were incubated for an additional 5 h. Protein extracts were prepared, separated by SDS-PAGE, transferred to PVDF membrane, and subjected to Western blot analysis with antibodies against XIAP, hnRNP A1, and GAPDH.
We next examined whether the decrease in XIAP protein levels seen during osmotic shock is dependent on hnRNP A1. Cells were transfected with hnRNP A1 siRNA or a nonsilencing control siRNA, and 72 h later the growth media was replaced with growth medium containing sorbitol (0.6 M OSM) or nonsupplemented growth medium. After a 5-h incubation, protein extracts were harvested, and XIAP protein levels were determined by Western blot analysis. As we had previously observed, osmotic shock causes a decrease in XIAP levels (Figure 5C, lane 2). Importantly, the decrease in XIAP levels in response to osmotic shock is dependent on hnRNP A1, because transient knockdown of hnRNP A1 prevents the decrease in XIAP levels caused by osmotic shock (Figure 5C, compare lanes 2 and 4). Taken together, these findings indicate that shuttling of hnRNP A1 from the nucleus to the cytoplasm during cellular stress (e.g., osmotic shock) causes a decrease in XIAP protein levels, likely through the ability of cytoplasmic hnRNP A1 to reduce XIAP IRES-dependent translation.
DISCUSSION
We have undertaken a search for ITAFs that are involved in the function of the XIAP IRES and have identified hnRNP A1 as a novel XIAP ITAF. We show here that hnRNP A1 interacts with the XIAP IRES RNA sequence in vitro and is associated with XIAP mRNA in vivo. Importantly, cytoplasmic relocalization of hnRNP A1, an event that is controlled by posttranslational modification of the protein during cellular stress conditions (e.g., osmotic shock), causes a significant decrease in XIAP IRES activity and a concomitant decrease in the level of endogenous XIAP protein. This perturbation of hnRNP A1 does not affect the integrity of mRNA containing the XIAP IRES, confirming that the effect of hnRNP A1 on XIAP IRES activity occurs at the translational level. On the basis of our findings, we conclude that cytoplasmic hnRNP A1 is a negative regulator of XIAP IRES-dependent translation, demonstrating a novel function for the cytoplasmic form of hnRNP A1. Moreover, our findings support the hypothesis that the subcellular compartmentalization of ITAFs controls their activity.
hnRNP A1 was recently shown to interact with the FGF-2 IRES, causing an upregulation of FGF-2 IRES activity (Bonnal et al., 2005). This is in contrast to the negative regulation of XIAP IRES activity that we have observed for hnRNP A1. However, we have also observed that hnRNP A1 has a negative effect on the activities of the c-myc, BiP, and VEGF IRES elements in an in vitro translation assay (Bonnal et al., 2005). These findings suggest that hnRNP A1 may be a dual-function ITAF, although we do not yet understand how hnRNP A1 can both positively regulate some IRES elements and negatively regulate others. As a negative regulator of IRES, hnRNP A1 may block binding sites for potent inducers of IRES activity. Indeed, we show that hnRNP A1 binds within the XIAP IRES core RNP-binding site (Figure 2), where we have previously localized binding of the La autoantigen, a positive regulator of XIAP IRES activity (Holcik and Korneluk, 2000). However, we also find that hnRNP A1 does not effectively compete with the La autoantigen for binding to XIAP IRES RNA in an in vitro UV cross-linking assay (Supplementary Figure S2), suggesting that displacement of the La autoantigen from the XIAP IRES is not the mechanism by which hnRNP A1 reduces XIAP IRES activity. This observation does not rule out the possibility that hnRNP A1 disrupts the binding of an unidentified ITAF within the core RNP-binding site of the XIAP IRES. Alternatively, hnRNP A1 may bind to some IRES (such as XIAP) and modify its structure in such a way that renders it nonfunctional, whereas hnRNP A1 may provide critical structural remodeling of other IRES (such as FGF-2) to induce a conformation that is amenable to ribosome recruitment.
XIAP is the prototype member of the family of intrinsic inhibitor of apoptosis (IAP) proteins. XIAP is critically involved in a number of cellular functions including direct caspase binding and inhibition, modulation of receptor-mediated signal transduction, and protein ubiquitination (reviewed in Liston et al., 2003). IRES-dependent translation permits de novo synthesis of XIAP during conditions that inhibit cap-dependent translation (Holcik et al., 1999), and therefore allows XIAP expression to persist during the commitment phase of apoptosis. The decision whether to commit to death or not is dependent on the abundance and activity of XIAP, which is partially determined by the level of XIAP IRES activity and is therefore linked to the function of XIAP ITAFs, such as hnRNP A1. Under proapoptotic conditions XIAP activity must be removed to allow apoptosis to proceed. Pre-existing XIAP can be neutralized by binding of Smac/DIABLO, XAF1, or HtrA1/Omi to XIAP (Du et al., 2000; Verhagen et al., 2000; Liston et al., 2001; Suzuki et al., 2001). However, because of persistent IRES activity, new XIAP protein will continue to be produced, and therefore the abundance of XIAP protein may exceed the capacity of XAF1, Smac/DIABLO, and HtrA1/Omi to regulate XIAP activity. An hnRNP A1–mediated reduction in XIAP IRES function would serve to shut off de novo XIAP synthesis, and combined with the neutralization of pre-existing XIAP by XAF1, Smac/DIABLO, and HtrA2/Omi, would permit execution of the cell death program. On the basis of our observations, we hypothesize that hnRNP A1 is redistributed from the nucleus to the cytoplasm during cellular stress and binds to the XIAP IRES to reduce XIAP translation when cell death is favorable.
hnRNP A1 has been shown to undergo posttranslational modification during cellular stress and apoptosis, which may affect its ability to modulate XIAP IRES activity. During osmotic shock, a cellular stress that results in apoptosis (Qin et al., 1997; Edwards et al., 1998; Galvez et al., 2001), hnRNP A1 has been shown to accumulate in the cytoplasm (van der Houven van Oordt et al., 2000). This redistribution of hnRNP A1 is dependent on the phosphorylation of hnRNP A1 at several serine residues at the carboxy terminus of the protein (known as the F-peptide), which blocks Trn1-dependent nuclear import of hnRNP A1 (Allemand et al., 2005). In this case, osmotic stress causes the redistribution of hnRNP A1 to the cytoplasm where it can bind to the XIAP IRES to reduce XIAP translation. In support of this hypothesis we show that a cytoplasmically restricted mutant of hnRNP A1 significantly reduces XIAP IRES activity (Figure 3B) and causes a greater reduction in endogenous XIAP proteins levels than the primarily nuclear wild-type hnRNP A1 (Figure 4A). Moreover, exposure of cells to high-osmolarity growth medium reduces XIAP expression, and this reduction in XIAP protein levels can be abrogated by transient knockdown of hnRNP A1 (Figure 5C), indicating that the reduction of XIAP expression in response to osmotic shock is dependent on hnRNP A1. It is also important to note that because hnRNP A1 is normally present in the nucleus (Pinol-Roma and Dreyfuss, 1992), depletion of hnRNP A1 under normal growth conditions would be predicted to have only a modest effect on IRES-dependent translation of XIAP, which we in fact observed (Figure 5C and data not shown). A recent report has shown that treatment of cells with sodium butyrate causes redistribution of hnRNP A1 from the nucleus to the cytoplasm (Tan et al., 2006). Importantly, sodium butyrate treatment also results in a reduction in XIAP protein levels and sensitization of human glioma cells to tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)-mediated apoptosis (Kim et al., 2005). We have observed that overexpression of a cytoplasmically localized mutant of hnRNP A1 significantly reduces XIAP IRES activity and decreases XIAP protein levels (Figures 3B and 4A), raising the possibility that sodium butyrate treatment sensitizes cells to apoptosis through the redistribution of hnRNP A1 from the nucleus to the cytoplasm, where hnRNP A1 significantly decreases IRES-dependent translation of XIAP.
Our findings suggest that a critical event for hnRNP A1-dependent regulation of XIAP translation is the relocalization of hnRNP A1 itself. Given that many other ITAFs are known to shuttle between the nucleus and the cytoplasm, we propose that the subcellular localization of ITAFs is critical for their role as regulators of IRES-dependent translation. Indeed, the c-myc ITAF hnRNP C1/C2 was found to translocate from the nucleus to the cytoplasm during the G2/M phase of the cell cycle, resulting in a concomitant increase in IRES-dependent translation of c-myc protein (Kim et al., 2003). Recent reports have described hnRNP A1 posttranslational modification in response stress stimuli (Allemand et al., 2005), and these posttranslational modifications are dependent on the activation of signaling pathways (van der Houven van Oordt et al., 2000). Future experiments delineating the signaling pathways that control hnRNP A1 modulation of XIAP IRES activity will increase our understanding of how XIAP IRES activity, and consequently cellular survival during stress, is regulated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Adrian Krainer for plasmid constructs, Dr. Robert Screaton for critical comments on the manuscript and useful suggestions, and members of our laboratory for helpful discussions. Work in the laboratory of M.H. was supported by grants from the Canadian Institutes of Health Research (CIHR MOP 43984), Premier's Research Excellence Award, Canada Foundation for Innovation, and Ontario Research and Development Challenge Fund. Work in the laboratory of S.V. was supported by INSERM, Fondation de France, and FRM (Equipe FRM, soutenue par la Fondation Recherche Médicale). M.H. is a CIHR New Investigator.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0515) on February 7, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Allemand E., Guil S., Myers M., Moscat J., Caceres J. F., Krainer A. R. Regulation of heterogeneous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A. C., Vagner S. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 2005;280:4144–4153. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- Chan E. K., Tan E. M. Human autoantibody-reactive epitopes of SS-B/La are highly conserved in comparison with epitopes recognized by murine monoclonal antibodies. J. Exp. Med. 1987;166:1627–1640. doi: 10.1084/jem.166.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Fang M., Li Y., Li L., Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Edwards Y. S., Sutherland L. M., Power J. H., Nicholas T. E., Murray A. W. Osmotic stress induces both secretion and apoptosis in rat alveolar type II cells. Am. J. Physiol. 1998;275:L670–L678. doi: 10.1152/ajplung.1998.275.4.L670. [DOI] [PubMed] [Google Scholar]
- Galvez A., Morales M. P., Eltit J. M., Ocaranza P., Carrasco L., Campos X., Sapag-Hagar M., Diaz-Araya G., Lavandero S. A rapid and strong apoptotic process is triggered by hyperosmotic stress in cultured rat cardiac myocytes. Cell Tissue Res. 2001;304:279–285. doi: 10.1007/s004410100358. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Hellen C. U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Holcik M., Gordon B. W., Korneluk R. G. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Graber T., Lewis S. M., Lefebvre C. A., Lacasse E., Baird S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the βgal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Korneluk R. G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Lefebvre C., Yeh C., Chow T., Korneluk R. G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell. Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N., Korneluk R. G. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/s0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- Kapp L. D., Lorsch J. R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kim E. H., Kim H. S., Kim S. U., Noh E. J., Lee J. S., Choi K. S. Sodium butyrate sensitizes human glioma cells to TRAIL-mediated apoptosis through inhibition of Cdc2 and the subsequent downregulation of survivin and XIAP. Oncogene. 2005;24:6877–6889. doi: 10.1038/sj.onc.1208851. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Paek K. Y., Choi K., Kim T. D., Hahm B., Kim K. T., Jang S. K. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 2003;23:708–720. doi: 10.1128/MCB.23.2.708-720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Paek K. Y., Ha S. H., Cho S., Choi K., Kim C. S., Ryu S. H., Jang S. K. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 2004;24:7878–7890. doi: 10.1128/MCB.24.18.7878-7890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., Back S. H., Rho J., Lee S. H., Jang S. K. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 2001;29:5009–5016. doi: 10.1093/nar/29.24.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann M., Gopfert U., Siewe B., Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. M., Holcik M. IRES in distress: translational regulation of the inhibitor of apoptosis proteins XIAP and HIAP2 during cell stress. Cell Death Differ. 2005;12:547–553. doi: 10.1038/sj.cdd.4401602. [DOI] [PubMed] [Google Scholar]
- Liston P., Fong W. G., Kelly N. L., Toji S., Miyazaki T., Conte D., Tamai K., Craig C. G., McBurney M. W., Korneluk R. G. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat. Cell Biol. 2001;3:128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- Liston P., Fong W. G., Korneluk R. G. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- MacGregor G. R., Nolan G. P., Fiering S., Roederer M., Herzenberg L. A. Use of the E. coli lacZ (β-galactosidase) as a reporter gene. In: Murray E. J., Walker J. M., editors. Methods in Molecular Biology. Vol. 7. Clifton, NJ: Humana Press; 1991. pp. 217–235. [DOI] [PubMed] [Google Scholar]
- Marissen W. E., Lloyd R. E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mitchell S. A., Brown E. C., Coldwell M. J., Jackson R. J., Willis A. E. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M. A. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Qin S., Minami Y., Kurosaki T., Yamamura H. Distinctive functions of Syk and Lyn in mediating osmotic stress- and ultraviolet C irradiation-induced apoptosis in chicken B cells. J. Biol. Chem. 1997;272:17994–17999. doi: 10.1074/jbc.272.29.17994. [DOI] [PubMed] [Google Scholar]
- Spriggs K. A., Bushell M., Mitchell S. A., Willis A. E. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Imai Y., Nakayama H., Takahashi K., Takio K., Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Tan H. T., Zubaidah R. M., Tan S., Hooi S. C., Chung M. C. 2-D DIGE analysis of butyrate-treated HCT-116 cells after enrichment with heparin affinity chromatography. J. Proteome Res. 2006;5:1098–1106. doi: 10.1021/pr050435r. [DOI] [PubMed] [Google Scholar]
- Vagner S., Gensac M. C., Maret A., Bayard F., Amalric F., Prats H., Prats A. C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Houven van Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., Caceres J. F. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen A. M., Ekert P. G., Pakusch M., Silke J., Connolly L. M., Reid G. E., Moritz R. L., Simpson R. J., Vaux D. L. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Warnakulasuriyarachchi D., Cerquozzi S., Cheung H. H., Holcik M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 2004;279:17148–17157. doi: 10.1074/jbc.M308737200. [DOI] [PubMed] [Google Scholar]
- Yoon A., Peng G., Brandenburger Y., Zollo O., Xu W., Rego E., Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.