Abstract
Tight control of translation is fundamental for eukaryotic cells, and deregulation of proteins implicated contributes to numerous human diseases. The neurodegenerative disorder spinocerebellar ataxia type 2 is caused by a trinucleotide expansion in the SCA2 gene encoding a lengthened polyglutamine stretch in the gene product ataxin-2, which seems to be implicated in cellular RNA-processing pathways and translational regulation. Here, we substantiate a function of ataxin-2 in such pathways by demonstrating that ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6, a component of P-bodies and stress granules, representing cellular structures of mRNA triage. We discovered that altered ataxin-2 levels interfere with the assembly of stress granules and cellular P-body structures. Moreover, ataxin-2 regulates the intracellular concentration of its interaction partner, the poly(A)-binding protein, another stress granule component and a key factor for translational control. Thus, our data imply that the cellular ataxin-2 concentration is important for the assembly of stress granules and P-bodies, which are main compartments for regulating and controlling mRNA degradation, stability, and translation.
INTRODUCTION
An essential step in the control of global gene expression in eukaryotes is the regulation of mRNA translation and degradation. Shortening of the poly(A) tail is the initial step to trigger mRNA for decay in two major mRNA degradation pathways in eukaryotic cells (Bernstein and Ross, 1989; Peltz et al., 1991; Decker and Parker, 1994; Beelman and Parker, 1995). In one pathway, the deadenylated mRNA is degraded by a cytoplasmic protein complex, the exosome, consisting of a number of exonucleases with 3′-5′ activity (Butler, 2002). In the other pathway, shortening of the poly(A) tail leads to the removal of the cap structure of the mRNA by the decapping proteins DCP1 and DCP2 facilitating 5′-3′ degradation of the mRNA through the exoribonuclease XRN1 (Beelman and Parker, 1995; Butler, 2002). These proteins colocalize in discrete cytoplasmic foci in mammalian cells, termed processing bodies or P-bodies (also known as DCP1- or GW182-bodies), indicating that mRNA decay is restricted to distinct cytoplasmic compartments in mammalian cells (van Dijk et al., 2002; Cougot et al., 2004). The human protein CCR4, which is involved in mRNA deadenylation, the decapping stimulating LSm proteins LSm1–7, the DEAD/H-box RNA helicase DDX6 (also known as RCK/p54), GW182, and Ge-1 are components of P-bodies (Bouveret et al., 2000; Eystathioy et al., 2002; Ingelfinger et al., 2002; Lykke-Andersen, 2002; Cougot et al., 2004; Yu et al., 2005). Moreover, the RNA-associated protein 55, hEDC3, Hedls as well as factors of the RISC complex localize to P-bodies in mammalian cells (Fenger-Gron et al., 2005; Liu et al., 2005; Sen and Blau, 2005; Yang et al., 2006). P-bodies represent dynamic structures that are in an equilibrium with polysomes and contain nontranslating mRNA (Teixeira et al., 2005). Remarkably, recent work demonstrated that mRNA molecules entering P-bodies can also be stored and return to a translational active state in yeast (Brengues et al., 2005).
Interestingly, under conditions of certain environmental stresses, P-bodies can localize to cytoplasmic stress granules (SGs) (Kedersha et al., 2005; Wilczynska et al., 2005). These cellular structures represent other ribonucleoprotein granules that seem to be in a dynamic equilibrium with polysomes as well and assemble and disassemble very rapidly in the cytoplasm of plant and mammalian cells after subjected to environmental stresses (Anderson and Kedersha, 2006). During these stress conditions, the translation of housekeeping genes is arrested and untranslated mRNA accumulates in SGs. One model for SG assembly is that the arrest of translation is initiated by activation of one or more eukaryotic initiation factor (eIF)2α kinases, which phosphorylate the translation initiation factor eIF2α. This causes a decline of the ternary complex eIF2-GTP-tRNAMet and an accumulation of the 48S* preinitiation complex resulting in the disassembly of polysomes and the assembly of SGs (Anderson and Kedersha, 2002a,b; Kedersha and Anderson, 2002). However, recently eIF2α phosphorylation-independent pathways, which target translation initiation, have been reported (Dang et al., 2006; Mazroui et al., 2006). Interference with the activity of another initiation factor, eIF4A, induces assembly of SGs. Moreover, inhibition of cap-dependent translation after poliovirus infection or treatment of cells with pateamine A leads to the assembly of SGs as well. SGs contain several ribonucleoproteins and a number of mRNA-stabilizing and -destabilizing factors such as elongation initiation factors, ribosomal subunits, RNA binding proteins, RNA stability factors, the poly(A)-binding protein (PABP) as well as the proteins TIA-1 and TIAR, which are essential for SG assembly (Anderson and Kedersha, 2006). Recently, the RasGAP-associated endoribonuclease G3BP and the translational regulator CPEB have been shown to be part of SGs and induce the assembly of SGs if overexpressed (Tourriere et al., 2003; Wilczynska et al., 2005). Interestingly, the survival motor neuron protein involved in the disorder spinal muscular atrophy and ataxin-2 (ATXN2) involved in the neurodegenerative disorder spinocerebellar ataxia type 2 (SCA2) are components of these cellular structures as well (Hua and Zhou, 2004; Ralser et al., 2005a).
The disorder SCA2 belongs to the polyglutamine disorder family, which includes Huntington's disease, spinobulbar muscular atrophy, dentatorubral pallidoluysian atrophy, and spinocerebellar ataxia types 1, 3, 6, 7, and 17. All of these disorders penetrate in midlife and exhibit a fatal disease progression over 10–20 yrs. On the molecular level, this family is characterized by an expansion of CAG repeats in the respective genes encoding an enlarged polyglutamine region in the disease-causing proteins (Tobin and Signer, 2000; Stevanin et al., 2002; Schols et al., 2004). Moreover, the molecular mechanisms contributing to most of these disorders and particularly SCA2 are far from being understood. However, evidence exists that an accumulation of ATXN2, as observed in SCA2 brains, might contribute to cellular dysfunction and SCA2 pathogenesis (Huynh et al., 1999; Koyano et al., 1999).
The biological function of ATXN2 is unknown, but evidence has been provided that ATXN2 is involved in the cellular RNA metabolism. Initially, ATXN2 was identified to interact with the protein A2BP1 (ATXN2 binding protein 1) that contains RNA recognition motifs (Shibata et al., 2000). Along this line, it has been reported that ATXN2 contains an Like Sm (LSm) domain that might be capable of RNA binding as observed for other LSm domain proteins (Neuwald and Koonin, 1998; Achsel et al., 2001; Albrecht et al., 2004). Further evidence for a likely role in RNA metabolism was provided by demonstrating that ATXN2 assembles with polysomes and interacts with the cytoplasmic poly(A)-binding protein 1 (PABP-C1) that functions in translation initiation and mRNA decay regulation (Ralser et al., 2005a; Satterfield and Pallanck, 2006). Proteomics-based results demonstrated that PABP and T-plastin, which itself associates with ATXN2, are present together in large protein complexes mostly consisting of proteins involved in RNA processing (Ho et al., 2002; Blagoev et al., 2003). Interestingly, the expression of ATXN2 results in enormous growth defects in yeast deficient for the actin-bundling protein fimbrin, the functional yeast orthologue of the mammalian plastin proteins (Ralser et al., 2005c). Regarding this issue, it is remarkable that the Drosophila melanogaster ATXN2 homologue is a dosage-sensitive regulator of cytoskeletal actin filament formation by controlling translation, stability, or localization of mRNA encoding proteins involved in actin polymerization (Satterfield et al., 2002). Furthermore, other ATXN2 homologues have been implicated in various RNA-processing tasks as well. In Caenorhabditis elegans, the ATXN2 homologue is found in complexes with PAB1 and may be implicated in translational regulation (Ciosk et al., 2004). Additionally, the yeast homologue of ATXN2, named Pbp1 (Pab1-binding protein 1, also known as MRS16) is involved in important RNA-processing pathways such as RNA editing, pre-mRNA splicing, mRNA export, and degradation, which is clearly demonstrated by its comprehensive protein interaction network (Ralser et al., 2005a). Because ATXN2 and Pbp1 are functionally related as shown previously (Ralser et al., 2005a), we were questioning whether additional protein interactions identified in the yeast Pbp1 interaction network can be assigned to the human system. Here, we addressed the question whether the interaction between Pbp1 and Dhh1, which itself is a component of P-bodies and plays a major role in mRNA turnover and degradation (Coller et al., 2001; Fischer and Weis, 2002; Tseng-Rogenski et al., 2003), is evolutionary conserved in humans.
MATERIALS AND METHODS
Plasmids
For the generation of the plasmid pBTM-ATXN2-LSm/LSmAD, a DNA fragment encoding amino acids 254–475 of ATXN2 was amplified via polymerase chain reaction (PCR) from the plasmid p416GPD-ATX2 (Ralser et al., 2005a) by using the oligonucleotides ATXN2-Lsm-s-SalI (GAGTCGACTACGATTTCTTTTGATGGAAT) and ATXN2-Lsm-as-NotI (TATAGCGGCCGCTCTCTGAACTGCTGTGTATT), purified and subcloned into the SalI/NotI sites of the yeast two-hybrid bait vector pBTM117c. Plasmids pBTM-ATX2-NTQ22, pBTM-ATX2-NTQ79, pBTM-ATX2-FC (Ralser et al., 2005c), or pBTM-ATX2-FD (Ralser et al., 2005a) or pBTM-htt (Ralser et al., 2005b) were described previously. For the expression in mammalian cells, plasmids encoding MYC- or FLAG-tagged versions of the LSm/LSmAD domain of ATXN2 were generated by isolating the respective DNA fragment from plasmid pGAD-ATXN2-LSm/LSmAD by treatment with SalI/NotI and ligation into the vector pCMV-MYC (SalI/NotI; Clontech, Mountain View, CA) or pTL-FLAG-C (XhoI/NotI), respectively. For the generation of plasmid pCMV-MYC-ATXN2-Q22, plasmid p416GPD-ATX2 was treated with XbaI, followed by blunting with T4 DNA polymerase and subsequent SalI digestion. Then, the resultant DNA fragment was purified and subcloned into pCMV-MYC, which has been treated with EcoRI, T4 DNA polymerase and subsequently with SalI. Plasmid pCMV-MYC-ATXN2-Q79 was generated by subcloning a DNA fragment containing the respective CAG expansion isolated from the plasmid p423GALL-ATXN2-Q79 by treatment with AscI and KpnI (Ralser et al., 2005c) and ligated into the AscI/KpnI sites of pCMV-MYC-ATXN2-Q22.
For the generation of plasmids encoding full-length human DDX6 (NM_004397), the corresponding cDNA was amplified from a human fetal cDNA library (Clontech) by using the oligonucleotide pair DDX6-CDS-s-SalI (GAGTCGACAATGGGTCTGTCCAGTCAAAA) and DDX6-as-NotI (TATAGCGGCCGCTGTCAAAGCATGCTTGTT). The resultant DNA fragment was treated with the restriction enzymes SalI and NotI and ligated into the SalI/NotI sites of the yeast two-hybrid prey plasmid pACT4–1b or into the XhoI/NotI sites of the mammalian expression vector pTL-FLAG-C. Underlined primer sequences represent restriction sites.
Yeast Two-Hybrid Analysis
For the directed yeast two-hybrid analysis, we transformed the yeast strain L40ccua (MATa his3Δ200 trp1-901 leu2-3,112 LYS2::(lexAop)4-HIS3 ura3:: (lexAop)8-lacZ ADE2::(lexAop)8-URA3 gal80 canR cyh2R) with the respective bait and prey plasmids as indicated. All bait proteins were tested for a potential autoactivation of the reporter genes in earlier studies (Ralser et al., 2005a,c; Ralser, unpublished data). Afterward, transformants were selected on synthetic complete media lacking amino acids tryptophan and leucine. Then, single colonies were isolated and spotted onto synthetic complete media lacking tryptophane, leucine, histidine, and uracil as well as onto nylon membranes. The activity of the reporter genes was analyzed by monitoring the growth of the respective transformants after incubation of plates for 3 d at 30°C. The activity of β-galactosidase was determined as described previously (Ralser et al., 2005a).
Cell Cultivation and Transfection
DU145, SH-SY5Y, and human embryonic kidney (HEK)293T cells were cultivated in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 100 U/ml penicillin/G-streptomycin (Biochrom, Berlin, Germany) and 10% heat-inactivated fetal bovine serum (Biochrom) at 37°C and 5% CO2. For overexpression studies, cells were grown until a confluence of 50–70%. Transfections were performed according to the recommendations of the manufacturer using Polyfect (QIAGEN, Hilden, Germany) and 1–2 μg of each expression plasmid. Afterward, transfected cells were incubated for 8–24 h to allow transient expression of proteins.
RNA Interference
For RNA interference experiments, we used different small interfering RNA (siRNA) molecules obtained from either QIAGEN (Hs_ATXN2_1_HP [siATXN2#1], Hs_ATXN2_2_HP[siATXN2#2], and Hs_ATXN2_3_HP[siATXN2#3]) or Invitrogen (ATXN2-HSS109492[siATXN2#4], ATXN2-HSS109493[siATXN2#5], and ATXN2-HSS109494[siATXN2#6]). Additionally, endoribonuclease-prepared siRNA (esiRNA) molecules were created by using the primers ATXN2 FW T7 siRNA (5′-CGTAATACGACTCACTATAGGGGGGTCATCAACAGCCAACTCCAG) and ATXN2 REV T7 siRNA (5′-CGTAATACGACTCACTATAGGGGGAGTATGTGGGTGCAGGGTAGC), which both contain T7 polymerase recognition sites (Henschel et al., 2004). As controls, the siRNA molecules nonsilencing (siControl#1) and Hs-C21orf66_1 (siControl#2) from QIAGEN were used. For transfection, 60,000–75,000 cells were plated in a 12-well plate and transfected with 300 ng of siRNA or esiRNA molecules by using Hiperfect reagent (QIAGEN) according to the manufacturer's recommendations if not indicated elsewhere. For the immunofluorescence studies, cells were plated on cover slips before transfection. After 68–72 h, cells were harvested and used for total RNA isolation and protein preparations, or they were further processed for immunofluorescence microscopy.
RNA Extraction and Reverse Transcription
Three days after transfection, total RNA was extracted from cultured HEK293T cells by using RNeasy mini kit (QIAGEN), following the manufacturer's instructions. All RNA samples were treated on-column with RNase-free DNase I, quantified by UV-spectrophotometry, and checked for integrity by gel electrophoresis. Afterward, reverse transcription reactions were performed with random hexamer primers and SuperScriptII reverse transcriptase (Invitrogen). In total, 1 μg of total RNA for each sample was converted into cDNA in a 20-μl reaction and diluted to 12.5 ng/μl equivalent total RNA.
Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
For the quantitative real-time RT-PCR analyses, the following predesigned, gene-specific TaqMan probes from Applied Biosystems (Foster City, CA) were used: Hs99999903_m1 for β-actin (ACTB), Hs00268077_m1 for ataxin-2 (ATXN2), and Hs00743792_s1 for PABP-C1. All assays met the amplification efficiency criteria of 100 ± 10% ([application note 127AP05-02]; www.appliedbiosystems.com) and were comparable with each other. Real-time RT-PCRs were performed in triplicate in 10 μl by using the TaqMan Universal PCR Master Mix (Applied Biosystems) and processed with the ABI Prism 7900HT sequence detection system (Applied Biosystems). The thermal cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s/60°C for 1 min. Amplification plot and predicted threshold cycle (Ct) values were obtained with the sequence Detection Software (SDS 2.1; Applied Biosystems). A common threshold value was chosen for all genes, and the baseline was set manually for individual genes.
For each target gene, the data were first normalized to the endogenous control gene (β-actin), and the ratios of siRNA-transfected cells/mock-transfected cells were calculated using comparative ΔΔCt method following the instructions of the User Bulletin #2 (Applied Biosystems). Then, the final ratios and standard deviations were calculated as the geometric mean of the ratios and its standard deviations of independent biological replicate experiments as indicated.
Coimmunoprecipitation
SH-SY5Y and HEK293T cells were washed in phosphate-buffered saline (PBS) and harvested by incubation with lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5% protease inhibitor (Complete tablets; Roche Diagnostics, Mannheim, Germany) and 25 U/ml benzonase (Merck, Darmstadt, Germany) for 30 min on ice. Afterward, the protein concentration of the cell lysates was determined. Five or 1 μl of the respective primary antibody (anti-ATXN2; BD Biosciences, San Jose, CA; anti-DDX6, Novus Biologicals, Littleton, CO) was added to each cell lysate (500 μg). After incubating the samples for 1 h at 4°C on a rotating wheel, 15 μl of IgG-conjugated M-280 Dynabeads (Dynal Biotech, Oslo, Norway) was added, and samples were incubated for additional 3 h at 4°C. Afterward, the Dynabeads were pulled down magnetically and washed three times with 3% bovine serum albumin/PBS followed by three washing steps with PBS. To elute the bound proteins from the beads, the samples were heated for 5 min at 95°C in SDS-sample buffer. Afterward, proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Whatman Schleicher and Schuell, Dassel, Germany) by using a PerfectBlue semidry electroblotter (PeqLAB Biotechnologie, Erlangen, Germany). Then, membranes were blocked in 5% milk solution for 1 h at room temperature or overnight at 4°C. If not indicated otherwise, membranes were incubated with the respective primary antibodies for 1 h at room temperature followed by incubation with the corresponding peroxidase (POD)-coupled secondary antibodies (1:5000, anti-mouse IgG POD conjugate; 1:5000, anti-rabbit IgG POD conjugate; Sigma Aldrich, Taufkirchen, Germany) or alkaline phosphatase (AP)-coupled secondary antibody (1:5000, anti-goat IgG AP conjugate). After treatment of the membranes with Western Lightning luminol reagent (PerkinElmer Life and Analytical Sciences, Boston, MA) or SuperSignal West Femto Solution (Pierce Chemical, Rockford, IL), the proteins were visualized by exposing the membranes to a BioMax XAR film (Eastman Kodak, Rochester, NY). In cases where AP-coupled antibodies were used, the membranes were incubated for 10 min with AP buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 5 mM MgCl2) and subsequently incubated with 10 ml of AP buffer containing 3.3 mg of nitrotetrazolium blue chloride (Sigma Aldrich) and 2.5 mg of 5-bromo-4-chloro-3-indolylphosphate (Sigma Aldrich).
Confocal Microscopy
For the various sets of experiments, the respective cell lines were plated on glass slides. Transfection of plasmids or siRNA molecules was performed as described above. Then, cells were fixed with 2% paraformaldehyde and treated with ice-cold methanol for 10 min and processed as described previously (Ralser et al., 2005c). Preparations were analyzed using a confocal microscope (LSM 510 META; Carl Zeiss, Jena, Germany) on an inverted stand (AxioVert 200M; Carl Zeiss) by using the objective Plan-NEOFLUAR 40× 1.3 oil differential interference contrast. Images were taken using Zeiss software LSM 5 version 3.5. The following antibodies were used: TIA-1 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), DDX6 (1:500; Novus Biologicals), hemagglutinin (1:500; Roche Diagnostics), FLAG (1:500; Sigma Chemie), DCP1 (1:600; van Dijk et al., 2002), PABP (1:500; (Kuyumcu-Martinez et al., 2004), ATXN2 (1:200; BD Biosciences); MYC (1:600; Upstate Biotechnology, Lake Placid, NY); α-goat-Cy3 or α-rabbit-Cy3 (1:500; Dianova, Hamburg, Germany), and α-mouse-fluorescein isothiocyanate (FITC) (1:500; Dianova).
RESULTS
DDX6 Interacts with the LSm/LSmAD Domain of ATXN2
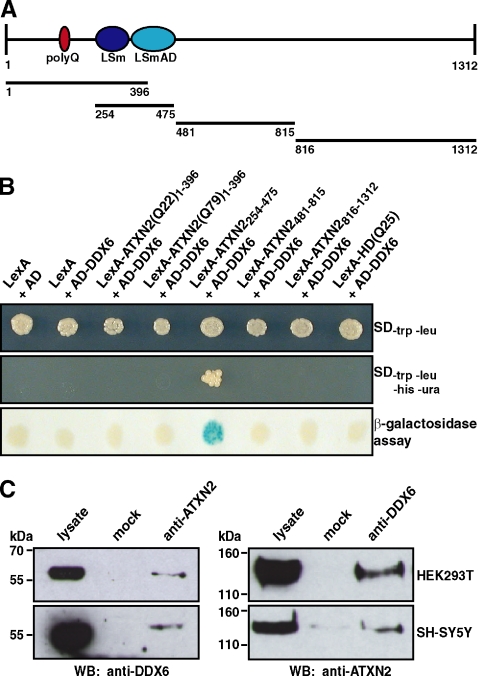
Recently, we have reported that ATXN2 and its yeast homologue Pbp1 are functionally conserved based on several observations described previously (Ralser et al., 2005a). First, overexpression of ATXN2 in yeast strains deleted for the magnesium transporter Mrs2 and mitochondrial introns resulted in the same phenotype as observed for Pbp1 overexpression. Moreover, the interaction between Pbp1 and the poly(A)-binding protein is conserved in mammalian cells, although both proteins have evolved different binding mechanisms (Ralser et al., 2005a). Thus, we investigated whether another interaction from the Pbp1 interactome is conserved in mammalian cells by focusing on the direct mammalian orthologue of the yeast Dhh1 protein, the DEAD/H-box RNA helicase DDX6. To test for a direct interaction, we generated a yeast two-hybrid prey plasmid encoding DDX6 and performed a yeast two-hybrid analysis. The yeast two-hybrid strain L40ccua was transformed with bait plasmids encoding different ATXN2 subregions, which cover the complete ATXN2 protein, as illustrated in Figure 1A, in combination with the prey plasmid encoding DDX6. We discovered that yeast cells coexpressing the fusion proteins AD-DDX6 and LexA-ATXN2254-475, which corresponds to the LSm/LSmAD domain of ATXN2, were viable on media lacking the respective amino acids as indicated, demonstrating that the respective reporter genes were activated (Figure 1B). Furthermore, a blue color shift was monitored in the membrane-based β-galactosidase assay, indicating that the lacZ reporter gene was transcribed as well. No reporter gene activities were monitored in yeast cells expressing fusion proteins LexA and AD, LexA and AD-DDX6 (controls), AD-DDX6 in combination with the other LexA-ATXN2 proteins, or AD-DDX6 and an unrelated bait protein LexA-HD(Q25). Thus, these experiments clearly demonstrated that ATXN2 and DDX6 interact in the yeast two-hybrid system.
Figure 1.
Interaction between ATXN2 and the DEAD/H-box RNA helicase DDX6. (A) Schematic illustration of ATXN2. Ellipses represent the polyQ-region (dark red), LSm and LSmAD domain (dark blue and light blue, respectively). Bars below display ATXN2 regions used in the directed yeast two-hybrid analysis. (B) Yeast two-hybrid analysis. Yeast strain L40ccua was transformed with the respective plasmids to coexpress the different LexA and AD fusion proteins as indicated. Afterward, transformants were isolated and spotted on selective media or on a membrane to analyze the activity of the reporter genes. (C) Coimmunoprecipitation. Cell lysates were prepared from HEK293T and SH-SY5Y cells. Coimmunoprecipitation was performed with 5 μl of α-ATXN2 antibody (left) or 1 μl of α-DDX6 antibody (right). Precipitated proteins were visualized with the antibodies indicated.
To verify this result in another experimental system, we performed coimmunoprecipitation experiments using different mammalian cell lines. Therefore, lysates from HEK293T and SH-SY5Y cells were prepared in the presence of an endonuclease to exclude that binding of both proteins is a bridging effect due to RNA or mediated by other interaction partners of ATXN2, which are known to bind RNA, as described in Materials and Methods. Subsequently, the lysates were incubated with antibodies directed against ATXN2 or DDX6. Afterward, the samples were supplemented with magnetic beads coupled to secondary antibodies and incubated at 4°C. Then, beads were pulled down magnetically and washed, and the eluted proteins were analyzed by immunoblotting. As shown in Figure 1C, endogenous DDX6 was precipitated from cell lysates prepared from HEK293T and SH-SY5Y by using an antibody directed against ATXN2. No DDX6 was precipitated from the control. In addition, endogenous ATXN2 was precipitated using an antibody directed against DDX6. In sum, this set of experiments showed that an association between ATXN2 and DDX6 occurs in mammalian cells. Thus, the interaction between the yeast proteins Pbp1 and Dhh1 is indeed conserved from yeast to humans.
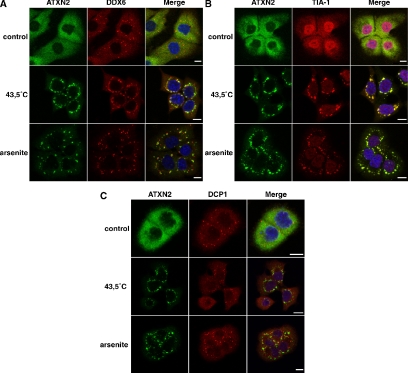
Whereas DDX6 localizes to distinct cytoplasmic foci representing P-bodies (Cougot et al., 2004; Wilczynska et al., 2005), ATXN2 seems to be evenly distributed throughout the cytoplasm of mammalian cells, for example, COS-1, HEK293T, and SH-SY5Y; no obvious accumulation of endogenous ATXN2 in particular cellular structures was observed (Ralser et al., 2005a,c; Figure 2A, control). Because an association of DDX6 and SGs has been demonstrated recently (Wilczynska et al., 2005), we further investigated whether DDX6 and ATXN2, which itself is a component of SGs (Ralser et al., 2005a), colocalize under stress conditions. We primarily used the cell line DU145, because DU145 cells were previously used for studying the assembly of SGs (Kedersha et al., 1999, 2000). To induce SGs, cells were treated with NaAsO2 or heat-shocked and then fixed. The cellular localization of endogenous ATXN2 and DDX6 was analyzed by confocal immunofluorescence microscopy. This analysis demonstrated that ATXN2 and DDX6 did colocalize in distinct cellular structures representing SGs (Figure 2A). Moreover, we also observed that SGs and P-bodies did overlap to some extent, however, they only overlapped at the edges of both structures, indicating recruitment of P-bodies to SGs as published previously (Kedersha et al., 2005; Wilczynska et al., 2005). To further confirm that the cellular structures ATXN2 and DDX6 localizes to are definitely SGs, we performed additional colocalization studies including the TIA-1 protein, because this protein is a well-accepted marker protein for SGs (Kedersha et al., 1999; Anderson and Kedersha, 2006). As expected, ATXN2 and TIA-1 did colocalize in distinct cytoplasmic foci in cells after treatment with arsenite or heat (Figure 2B). Interestingly, all cytoplasmic foci stained by the TIA-1 antibody were ATXN2 positive, indicating that ATXN2 represents an equal marker for SGs. Moreover, we included the DCP1 protein, which represents a marker protein for P-bodies. In contrast to DDX6, DCP1 was not present in SGs as published previously (Cougot et al., 2004). No colocalization of DCP1 and ATXN2 was observed in heat- or arsenite-treated cells (Figure 2C), revealing that the localization of ATXN2, like for TIA-1, is restricted to SGs.
Figure 2.
ATXN2 and DDX6 colocalize in stress granules. DU145 cells were treated with 0.5 mM arsenite or heat shocked at 43.5°C for 1 h. Control cells were left untreated at 37°C. Colocalization of endogenous ATXN2 and DDX6 (A), TIA-1 (B), or DCP1 (C). Nuclei were stained with Hoechst. Bars, 10 μm.
Elevated ATXN2 Levels Interfere with Cellular P-Body Structures
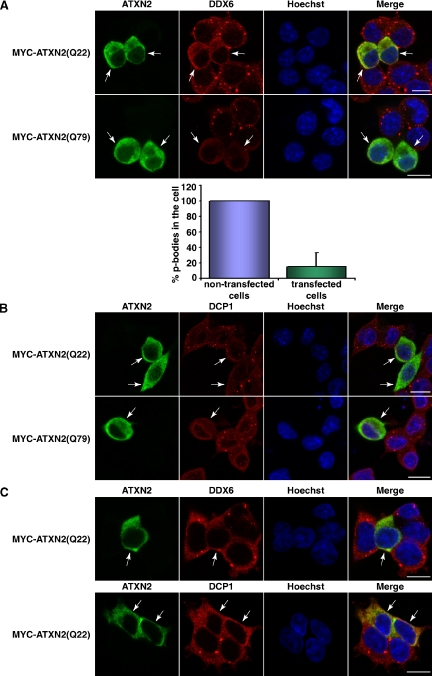
In postmortem brains of SCA2 patients, possessing an expansion of the CAG repeat in the SCA2 gene encoding a lengthened polyglutamine stretch within the ATXN2 protein, an accumulation of ATXN2 has been detected. Moreover, elevated ATXN2 levels sensitize certain neuroblastoma cells for apoptosis (Huynh et al., 1999; Koyano et al., 1999; Wiedemeyer et al., 2003). Therefore, we set out to directly investigate whether an elevated level of normal or disease-causing expanded ATXN2 protein would have an impact on the cellular P-body structures. To accomplish this task, we used the neuroblastoma cell line SH-SY5Y for this type of analyses and plasmids encoding full-length MYC-tagged ATXN2 with 22 (normal repeat) or 79 (pathogenic repeat) consecutive glutamines. Twenty-four hours after transfection, cells were fixed and stained for overexpressed ATXN2 and for endogenous DDX6. SH-SY5Y cells overexpressing ATXN2 with 22 or 79 consecutive glutamines exhibited an altered cellular distribution of DDX6 (Figure 3A). Notably, DDX6 was more diffusely distributed within the cytoplasm, and the number of P-bodies present was greatly reduced indicating an interference of high ATXN2 levels with P-body structures (Figure 3A, bottom). To confirm that the number of cellular P-bodies was indeed reduced under these conditions, we stained once more for another P-body marker protein DCP1. In agreement, we observed that SH-SY5Y cells transiently overexpressing ATXN2 proteins display an altered cellular distribution of DCP1 (Figure 3B), and again the number of P-bodies was reduced. Additionally, to substantiate these findings, we performed the same set of experiments in the cell lines HEK293T (Figure 3C) or DU145 (Figure 5A, control) and obtained comparable results.
Figure 3.
ATXN2 overexpression interferes with P-body assembly. (A) SH-SY5Y cells were transiently transfected with plasmids pCMV-MYC-ATXN2-Q22 or pCMV-MYC-ATXN2-Q79. For staining exogenous ATXN2 and endogenous DDX6, cells were incubated with antibodies directed against the MYC-tag and DDX6, followed by treatment with secondary antibodies coupled to FITC or Cy3, respectively. For quantitative analysis, the percentage of P-bodies in nontransfected versus transfected cells was calculated using the AxioVision software. Here, P-bodies of cells were counted in each picture taken and divided through the cell number. The mean value of P-bodies in the cells counted was calculated and SD was weighted. Then, the number of P-bodies counted for the nontransfected cells was set as 100%, and the number of P-bodies counted for the transfected cells was aligned. (B) Twenty-four hours after transfection, SH-SY5Y cells transiently overexpressing MYC-ATXN2(Q22) or MYC-ATXN2(Q79) or HEK293T transiently overexpressing MYC-ATXN2(Q22) (C) were stained for the MYC-tag and DDX6 or DCP1, respectively. Nuclei were stained with Hoechst. Bars, 10 μm. Arrows indicate transfected cells.
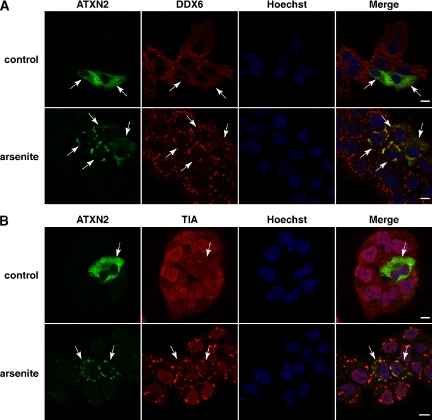
Figure 5.
Overexpression of ATXN2 does not influence SG formation. DU145 cells were transiently transfected with plasmid pCMV-MYC-ATXN2-Q22. Eight hours post transfection, cells were treated with 0.5 mM arsenite for 1 h. Control cells were left untreated at 37°C. For visualization of MYC-tagged ATXN2, cells were incubated with an antibody directed against the MYC-tag and antibodies against DDX6 (A) or TIA-1 (B) under normal and oxidative stress conditions as indicated. Arrows indicate transfected cells. Nuclei were stained with Hoechst. Bars, 10 μm.
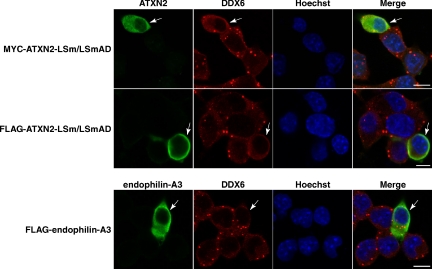
Because we expected that the cellular displacement of DDX6 could be based on a direct recruitment of DDX6, we overexpressed the LSm/LSmAD-domain of ATXN2, the domain demonstrated to bind DDX6 (Figure 1B), in SH-SY5Y cells. Interestingly, cells overexpressing this domain exhibited a delocalization of DDX6 (Figure 4). Finally, to confirm that the observed effects are specific due to an elevated ATXN2 level, we included the endophilin-A3 protein, another interaction partner of ATXN2 (Ralser et al., 2005c), in our studies. In this case, we did not observe any effect on the cellular P-body structures (Figure 4). Together, these experiments demonstrate that an elevated cellular ATXN2 level, as observed in SCA2 patients and some neuroblastomas (Huynh et al., 1999; Koyano et al., 1999; Wiedemeyer et al., 2003), can interfere with the presence of P-body structures.
Figure 4.
The LSm/LSmAD domain of ATXN2 is accountable for the interference with P-body structures. SH-SY5Y cells were transiently transfected with plasmid pCMV-MYC-ATXN2-LSm/LSmAD or pTL-FLAG-ATXN2-LSm/LSmAD. For staining exogenous ATXN2 and endogenous DDX6, cells were incubated with antibodies directed against the MYC- or FLAG-tag and DDX6, followed by treatment with secondary antibodies coupled to FITC or Cy3, respectively. To overexpress an unrelated protein as control, cells were transfected with plasmid pTL-FLAG-endophilin-A3. Nuclei were stained with Hoechst. Bars, 10 μm. Arrows indicate transfected cells.
SG Formation Does Not Seem to be Affected by an Elevated ATXN2 Concentration
Recently, a functional interplay between P-bodies and SGs has been described (Kedersha et al., 2005; Wilczynska et al., 2005). Because an elevated ATXN2 level seems to interfere with the presence of cellular P-body structures, we consequently addressed whether ATXN2 overexpression would impact the association of DDX6 with SGs as well. DU145 cells were transfected with plasmids encoding full-length MYC-tagged ATXN2 with 22 or 79 consecutive glutamines. After transfection, cells were treated with arsenite as described. Then, cells were fixed and stained for MYC-tagged ATXN2 and for endogenous DDX6. Association of DDX6 with SGs was detected to the same extent in cells exhibiting high levels of ATXN2 compared with untransfected cells (Figure 5A), indicating that an elevated ATXN2 level does not obviously interfere with the assembly of SGs. This observation was also supported by costaining ATXN2 and TIA-1 (Figure 5B). No obvious difference in SG number or size was apparent in transfected versus untransfected cells. In addition, cells transiently overexpressing ATXN2 with 79 consecutive glutamines did not display apparent effects on SG number or size as well (data not shown). However, we detected that the intracellular localization of TIA-1 in cells displaying high levels of ATXN2 with 22 or 79 consecutive glutamines was slightly altered (Figure 5B; data not shown); TIA-1 was not as prominent in the nucleus as it was in untransfected control cells. Thus, elevated ATXN2 levels did not seem to interfere with cellular SG structures under the chosen experimental conditions, but potentially affect the localization of TIA-1.
Reduced ATXN2 Levels Lead to Impaired SG Assembly
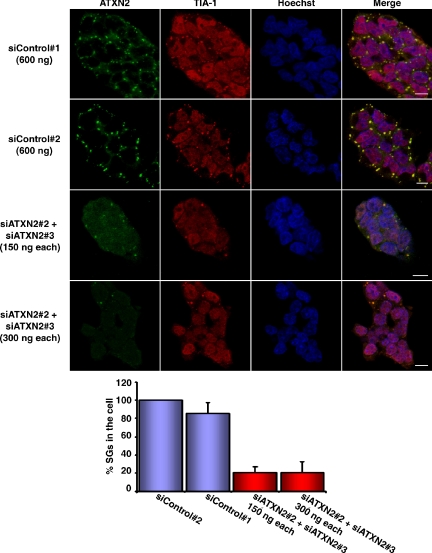
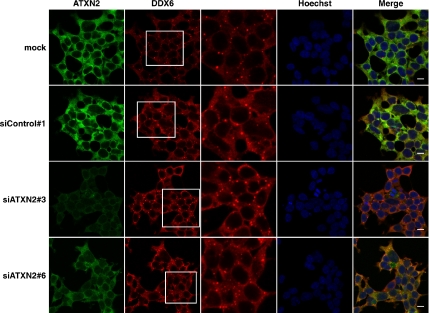
In the next step, we decided to perform RNA interference experiments to study the assembly of SGs in case the expression level of ATXN2 is reduced. In a first set of experiments, we analyzed for best knockdown efficiencies of ATXN2 performing quantitative real-time RT-PCR by using seven different siRNA molecules as described in Materials and Methods. Out of these seven siRNAs, three siRNA molecules (siATXN2#2, siATXN2#3, and siATXN2#6) and esiATXN2 resulted in a knockdown of ATXN2 of at least 65–75% in HEK293T cells. Because we were not able to reach same knockdown efficiencies using DU145 or SH-SY5Y cells, we chose HEK293T as cell line and the siRNA molecules siATXN2#2, siATXN2#3, siATXN2#6, and esiATXN2 for this type of experiments. Nonetheless, in an attempt to accomplish a better knockdown efficiency, we mixed the two siRNA molecules siATXN2#2 and siATXN2#3, which comprise different siRNA sequences. Indeed, knockdown efficiencies of ATXN2 greater than 75% were achieved when the two siRNA molecules were combined as analyzed by quantitative real-time RT-PCR (data not shown). Then, HEK293T cells were transfected with the different RNAi molecules and prepared for immunofluorescence microscopy. We observed that cells with lowered ATXN2 concentration exhibited a dramatic reduction in number and size of SG structures compared with control cells (Figure 6). Interestingly, TIA-1 and ATXN2 colocalized in all remaining SGs. Similar results were observed in case one single siRNA molecule was used for transfection; however, the effects were obvious but less prominent (data not shown). Moreover, we further investigated whether a reduced ATXN2 level would have an impact on P-body assembly as well. However, no obvious effect on P-body structures was observed in the presence of low ATXN2 concentration (Figure 7). Thus, ATXN2 seems to be required for the assembly of SGs.
Figure 6.
A reduced ATXN2 level prevents SG assembly. Top, HEK293T cells were cotransfected with a mix of siATXN2#2 and siATXN2#3 molecules. As controls, siControl#1 and siControl#2 were used as indicated. Sixty-eight hours post transfection, cells were incubated with 0.5 mM arsenite for 1 h. Nuclei were stained with Hoechst. Bars, 10 μm. Bottom, quantitative analysis. The percentage of TIA-1–positive SGs in control cells versus knockdown cells was determined using the AxioVision software. The number of cellular SGs was counted in each picture taken and divided through the cell number. The mean value of SGs in the cells counted was calculated and SD was weighted. Finally, the number of SGs counted for the control cells was set as 100%, and the number of SGs counted for the knockdown cells was aligned.
Figure 7.
A reduced ATXN2 level does not seem to affect P-body structures. HEK293T cells were transfected with siControl#1, siATXN2#3, or siATXN2#6 molecules or with transfection reagent only (mock). Endogenous levels of ATXN2 and DDX6 were visualized using antibodies against ATXN2 and DDX6. White boxes represent areas represented enlarged in the adjacent picture column. Nuclei were stained with Hoechst. Bars, 10 μm.
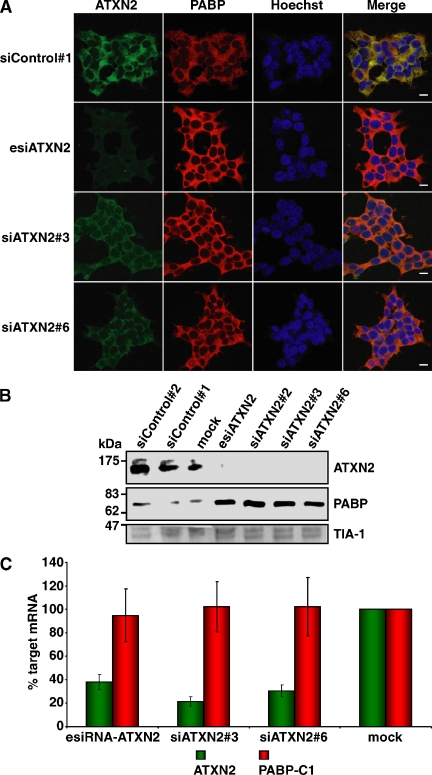
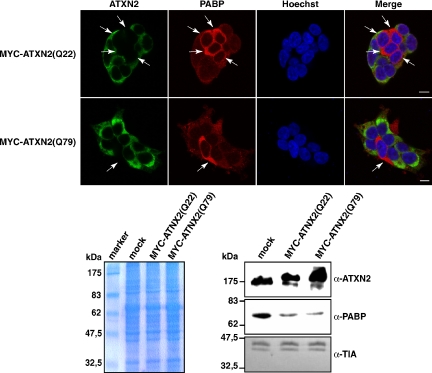
Furthermore, we included in the course of the above-described experiments the PABP protein as an additional component of SGs that is of particular interest, because of its interaction with ATXN2 (Ralser et al., 2005a). Strikingly, we observed that the PABP immunoreactivity was dramatically increased in all cells with diminished ATXN2 level (Figure 8A). To corroborate this remarkable finding, we performed immunoblot analysis by using HEK293T lysates (Figure 8B). Again, the level of PABP was elevated in cell lysates of the respective RNA interference samples compared with the corresponding control lysates. In addition, we investigated whether the increase in PABP concentration is occurring on transcriptional or translational level. Therefore, we performed quantitative real-time RT-PCR demonstrating that the changes in the intracellular PABP concentration are not caused by increased transcription of the PABP-C1 gene (Figure 8C). This result could give a hint in the direction that ATXN2 might destabilize PABP, based potentially on its interaction. Consequently, we transiently overexpressed ATXN2 with 22 or 79 glutamines and monitored the endogenous PABP level by immunofluorescence microscopy and by immunoblotting. Interestingly, we discovered that elevated levels of both ATXN2 proteins led to a decrease in the endogenous PABP concentration compared with untransfected cells (Figure 9). Thus, ATXN2 is a dosage-dependent regulator of its interaction partner PAPB.
Figure 8.
The endogenous PABP level is increased in the presence of a low intracellular ATXN2 level. (A) Knockdown studies. HEK293T cells were transfected with siControl#1, esiATXN2, siATXN2#3, or siATXN2#6. Endogenous levels of ATXN2 and PABP were visualized using antibodies against ATXN2 and PABP. (B) Western blot analysis. HEK293T cells transfected with siControl#1, siControl#2, siATXN2#2, siATXN2#3, siATXN2#6, esiATXN2, or transfection reagent (mock) were lysed. The same amount of each protein lysate was separated by SDS-PAGE and transferred to a nitrocellulose membrane. Antibodies directed against ATXN2, PABP, and TIA-1 were used for visualization of proteins. For presentation, the image was juxtaposed using Adobe Photoshop and Illustrator software (Adobe Systems, Mountain View, CA). (C) Quantitative real-time RT-PCR analysis of ATXN2 mRNA knockdown. Seventy-two hours after transfection, RNA from the treated cells was recovered and reverse transcribed. Target ATXN2 and PABP RNA levels were measured by quantitative real-time RT-PCR by using TaqMan assays. Values derived from quantitative real-time RT-PCR of control mock cells (transfected only with transfection reagent) were set as 100%, and the relative expression levels of cells transfected with ATXN2 siRNAs are indicated. Input cDNA in the different samples was normalized using real-time data for β-actin (ACTB). RNA levels are representative of five independent experiments. Error bars indicate SD.
Figure 9.
ATXN2 overexpression reduces the endogenous PAPB level. Top, HEK293T cells were transfected with plasmids encoding MYC-ATXN2(Q22) or MYC-ATXN2(Q79) proteins. Twenty-four hours post transfection, proteins were visualized with antibodies directed against the MYC-tag and PABP, respectively. Nuclei in all images presented were stained with Hoechst. Bars, 10 μm. Arrows indicate nontransfected cells. Bottom, Western blot analysis. HEK293T transiently overexpressing MYC-ATXN2(Q22) or MYC-ATXN2(Q79) proteins were lysed. The same amount of each protein lysate was separated by SDS-PAGE and transferred to a nitrocellulose membrane or stained with Coomassie to demonstrate equal loading of each lysate. Antibodies directed against ATXN2, PABP, and TIA-1 were used for visualization of proteins.
DISCUSSION
Previously, evidence has been provided that human ATXN2 plays a role in RNA-processing pathways (Shibata et al., 2000; Ralser et al., 2005a; Satterfield and Pallanck, 2006). Here, we present further data substantiating that ATXN2 takes part in important steps of mRNA control and regulation. We demonstrated that ATXN2 interacts via its LSm/LSmAD domain with the highly conserved DEAD/H-box RNA helicase DDX6, also termed RCK/p54, by using different experimental systems. Interestingly, this protein seems to be involved in mRNA decay due to its presence in so-called P-bodies (Coller et al., 2001; Fischer and Weis, 2002; Cougot et al., 2004). These are highly dynamic cellular structures that represent sites of mRNA decapping and degradation. Recently, it has been demonstrated that transiently overexpressed GFP-DDX6 or RFP-DDX6 is present in SGs as well, sustaining the dynamic transition of components shared by both cellular compartments under stress conditions (Wilczynska et al., 2005). Regarding this, we showed that ATXN2 and endogenous DDX6 colocalize within SGs in mammalian cells.
Subsequently, we addressed what the resulting functional consequences of alterations in the intracellular ATXN2 concentration on these cellular structures would be, because enhanced levels of ATXN2 have been observed in Purkinje cells of humans with age and in Purkinje cells of SCA2 patients, indicating that alterations of intracellular ATXN2 may contribute to cellular dysfunction and could correlate with disease progression (Huynh et al., 1999; Koyano et al., 1999). Interestingly, changes in the cellular concentration of ATXN2 seem to have an impact on cancer as well. Accumulation of ATXN2 sensitizes neuroblastoma cells of young children for apoptosis. Moreover, in neuroblastoma tumors, which contain an amplification of the MYCN gene, a significant lower ATXN2 concentration was observed in comparison with tumors without amplified MYCN (Wiedemeyer et al., 2003). In the first instance, we investigated whether increased intracellular levels of ATXN2 have an impact on P-body structures. We discovered that in almost all cells, which transiently overexpress ATXN2, the number of P-bodies was drastically reduced. This effect was observed overexpressing full-length ATXN2 with 22 or 79 glutamines and the individual LSm/LSmAD domain of ATXN2, the region responsible for binding DDX6. This result suggests that the delocalization of DDX6 is likely based on a recruitment mechanism. Along this line, knockdown experiments of DDX6 via RNAi technology demonstrated that the accumulation of P-body structures is severely affected in the absence of DDX6 showing that DDX6 is an essential component for the assembly of P-bodies (Andrei et al., 2005). Moreover, reduced levels of other P-body components such as LSm1, CCR4, or eIF4E-T result in failing of P-body accumulation (Andrei et al., 2005). Besides, an influence of reduced ATXN2 levels on P-body structures was not evident, indicating that in contrast to the mentioned core components, the presence of ATXN2 is not crucial for P-body assembly. Therefore, it might be plausible that high levels of ATXN2 would additionally interfere with the correct complexing of other LSm proteins, which are components of P-bodies as well, because most LSm domain proteins assemble as heteromers through their LSm domains (He and Parker, 2000). Interestingly, the Pbp1 interactome predicts an association of LSm proteins with ATXN2 (Ralser et al., 2005a).
In contrast to these observations, high levels of ATXN2 did not seem to interfere with the assembly of SGs under the chosen experimental conditions. However, TIA-1, which is essential for SG assembly (for review, see Anderson and Kedersha, 2002b) and one of its key components, displayed an altered localization in the respective cells. This finding is fairly interesting and clearly deserves further investigations. Moreover, we demonstrated that decreased ATXN2 levels impair the assembly of SGs. We noticed that mammalian cells with reduced ATXN2 levels displayed a higher PABP level compared with control cells. Conversely, cells overexpressing ATXN2 exhibited a lower PABP level. This finding is fairly significant, because PABP is one of the key proteins regulating mRNA translation and stability.
Alterations in the cellular level of PABP do not only affect synthesis of a variety of proteins, but the cellular protein expression profile as well (Ma et al., 2006). Overexpression of PABP leads to defects in cell division of the fission yeast Saccharomyces pombe (Tallada et al., 2002) and increases the translation termination efficiency in Saccharomyces cerevisiae (Cosson et al., 2002). Moreover, high PABP levels interfere with maturation-specific deadenylation and translational inactivation of maternal mRNAs in Xenopus oocytes (Wormington et al., 1996). In addition, elevated PABP levels have been detected in early stages of cancer supporting a link between cell growth and PABP levels (Verlaet et al., 2001). In contrast, depletion of PABP from a cell-free extract prevents the initiation of mRNA translation, because PABP mediates mRNA circularization through binding to the eIF4F translation initiation complex, which enhances translation by facilitating the recycling of ribosomes (Kahvejian et al., 2005). Interestingly, Yoshida and coworkers demonstrated that PABP depletion causes an additional reduction in the PABP-interacting protein 2 (PAIP2), demonstrating that PABP homeostasis is mediated by the stability of PAIP2 (Yoshida et al., 2006). One important player in this process seems to be the PABC-domain containing ubiquitin ligase HYD/EDD. In this light it is quite interesting that the PAM2 motif of ATXN2 shows in vitro affinity to the PABC domain of HYD as well (Lim et al., 2006b; Yoshida et al., 2006). However, we were unable to confirm an interaction of HYD-PABC and ATXN2 by using the yeast two-hybrid system (data not shown). Nonetheless, this fact could indicate that the interaction between HYD and ATXN2 might take place only under certain biological conditions, which yet need to be defined.
Because translational regulation is an important factor in regulating growth and differentiation, the deregulation of proteins implicated in these RNA-processing pathways has a great impact on the cellular homeostasis and consequently on several disorders. Therefore, it is quite intriguing that the experimental findings pinpoint ATXN2 function toward RNA homeostasis and translational control. Therefore, one might speculate that ATXN2 functions in the control and regulation of SG and P-body transition. In this light, one should keep in mind that ATXN2 has been implicated in transport processes, because it interacts with endophilin-A1 and -A3 (Ralser et al., 2005c), α-actinin 1, α-actinin 2, and secretogranin-1 (Lim et al., 2006a). Evidently, further studies are required to dissect the function of ATXN2 in translational regulation and how alterations in the intracellular ATXN2 level affect translational processes or potentially mRNA decay. On the one hand, one could speculate that ATXN2 interferes with SG and P-body structures by impairing the correct ribosome assembly granted by RNA helicases, which is required for the induction of SGs (Ripmaster et al., 1992; Mazroui et al., 2006). Moreover, one needs to elucidate in more detail the aspect whether the observed impairment of SG assembly in cells with reduced ATXN2 concentration could potentially arise due to alterations in the intracellular PAPB concentration as well, because PAPB is known to be a translational enhancer. Finally, it is of great interest to explore how disturbances of these RNA-processing and control pathways may contribute to the pathomechanisms underlying SCA2. Because we did not detect any differences between normal and mutant ATXN2 regarding their impact on SG and P-body homeostasis, it seems very likely that the disturbances underlying the disease state might be based to some extent on an increased cellular ATXN2 level.
ACKNOWLEDGMENTS
We are grateful to Richard E. Lloyd (Baylor College of Medicine, Houston, TX) and to Bertrand Séraphin (Centre de Genetique Moleculaire-Centre National de la Recherche Scientifique, Gif sur Yvette Cedex, France) for providing antibodies against PABP or DCP1, respectively. We are also grateful to Mario Albrecht (Max Planck Institute for Informatics, Saarbrücken, Germany) for helpful discussions. This work was funded by the Max Planck Society and the Federal Ministry of Education and Research (BMBF) in the framework of the National Genome Research Network (grant PRI-S02T03).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1120) on February 7, 2007.
REFERENCES
- Achsel T., Stark H., Luhrmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. USA. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M., Golatta M., Wullner U., Lengauer T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur. J. Biochem. 2004;271:3155–3170. doi: 10.1111/j.1432-1033.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Stressful initiations. J. Cell Sci. 2002a;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002b;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei M. A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C. A., Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Bernstein P., Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem. Sci. 1989;14:373–377. doi: 10.1016/0968-0004(89)90011-x. [DOI] [PubMed] [Google Scholar]
- Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. S. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- Ciosk R., DePalma M., Priess J. R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- Coller J. M., Tucker M., Sheth U., Valencia-Sanchez M. A., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., Philippe M., Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W. K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J. O. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem. Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Eystathioy T., Chan E. K., Tenenbaum S. A., Keene J. D., Griffith K., Fritzler M. J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Fischer N., Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 2000;12:346–350. doi: 10.1016/s0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- Henschel A., Buchholz F., Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113–W120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hua Y., Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572:69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Huynh D. P., Del Bigio M. R., Ho D. H., Pulst S. M. Expression of ataxin-2 in brains from normal individuals and patients with Alzheimer's disease and spinocerebellar ataxia 2. Ann. Neurol. 1999;45:232–241. doi: 10.1002/1531-8249(199902)45:2<232::aid-ana14>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D., Arndt-Jovin D. J., Luhrmann R., Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A., Svitkin Y. V., Sukarieh R., M'Boutchou M. N., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fitzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano S., Uchihara T., Fujigasaki H., Nakamura A., Yagishita S., Iwabuchi K. Neuronal intranuclear inclusions in spinocerebellar ataxia type 2, triple-labeling immunofluorescent study. Neurosci. Lett. 1999;273:117–120. doi: 10.1016/s0304-3940(99)00656-4. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez M., Belliot G., Sosnovtsev S. V., Chang K. O., Green K. Y., Lloyd R. E. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J. Virol. 2004;78:8172–8182. doi: 10.1128/JVI.78.15.8172-8182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006a;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Lim N. S., Kozlov G., Chang T. C., Groover O., Siddiqui N., Volpon L., De Crescenzo G., Shyu A. B., Gehring K. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J. Biol. Chem. 2006b;281:14376–14382. doi: 10.1074/jbc.M600307200. [DOI] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Musa T., Bag J. Reduced stability of mitogen-activated protein kinase kinase-2 mRNA and phosphorylation of poly(A)-binding protein (PABP) in cells overexpressing PABP. J. Biol. Chem. 2006;281:3145–3156. doi: 10.1074/jbc.M508937200. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F., Koonin E. V. Ataxin-2, global regulators of bacterial gene expression, and spliceosomal snRNP proteins share a conserved domain. J. Mol. Med. 1998;76:3–5. doi: 10.1007/s001090050184. [DOI] [PubMed] [Google Scholar]
- Peltz S. W., Brewer G., Bernstein P., Hart P. A., Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit. Rev. Eukaryot. Gene Expr. 1991;1:99–126. [PubMed] [Google Scholar]
- Ralser M., Albrecht M., Nonhoff U., Lengauer T., Lehrach H., Krobitsch S. An integrative approach to gain insights into the cellular function of human ataxin-2. J. Mol. Biol. 2005a;346:203–214. doi: 10.1016/j.jmb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Ralser M., Goehler H., Wanker E. E., Lehrach H., Krobitsch S. Generation of a yeast two-hybrid strain suitable for competitive protein binding analysis. Biotechniques. 2005b;39:165–166. 168. doi: 10.2144/05392BM01. [DOI] [PubMed] [Google Scholar]
- Ralser M., Nonhoff U., Albrecht M., Lengauer T., Wanker E. E., Lehrach H., Krobitsch S. Ataxin-2 and huntingtin interact with endophilin-A complexes to function in plastin-associated pathways. Hum. Mol. Genet. 2005c;14:2893–2909. doi: 10.1093/hmg/ddi321. [DOI] [PubMed] [Google Scholar]
- Ripmaster T. L., Vaughn G. P., Woolford J. L., Jr. A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl. Acad. Sci. USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield T. F., Jackson S. M., Pallanck L. J. A Drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics. 2002;162:1687–1702. doi: 10.1093/genetics/162.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield T. F., Pallanck L. J. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- Schols L., Bauer P., Schmidt T., Schulte T., Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- Sen G. L., Blau H. M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Shibata H., Huynh D. P., Pulst S. M. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- Stevanin G., Durr A., Brice A. Spinocerebellar ataxias caused by polyglutamine expansions. Adv. Exp. Med. Biol. 2002;516:47–77. doi: 10.1007/978-1-4615-0117-6_3. [DOI] [PubMed] [Google Scholar]
- Tallada V. A., Daga R. R., Palomeque C., Garzon A., Jimenez J. Genome-wide search of Schizosaccharomyces pombe genes causing overexpression-mediated cell cycle defects. Yeast. 2002;19:1139–1151. doi: 10.1002/yea.902. [DOI] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin A. J., Signer E. R. Huntington's disease: the challenge for cell biologists. Trends Cell Biol. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tseng-Rogenski S. S., Chong J. L., Thomas C. B., Enomoto S., Berman J., Chang T. H. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res. 2003;31:4995–5002. doi: 10.1093/nar/gkg712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B. Human Dcp 2, a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaet M., Deregowski V., Denis G., Humblet C., Stalmans M. T., Bours V., Castronovo V., Boniver J., Defresne M. P. Genetic imbalances in preleukemic thymuses. Biochem. Biophys. Res. Commun. 2001;283:12–18. doi: 10.1006/bbrc.2001.4731. [DOI] [PubMed] [Google Scholar]
- Wiedemeyer R., Westermann F., Wittke I., Nowock J., Schwab M. Ataxin-2 promotes apoptosis of human neuroblastoma cells. Oncogene. 2003;22:401–411. doi: 10.1038/sj.onc.1206150. [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Wormington M., Searfoss A. M., Hurney C. A. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- Yang W. H., Yu J. H., Gulick T., Bloch K. D., Bloch D. B. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Yang W. H., Gulick T., Bloch K. D., Bloch D. B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]