Abstract
Mammalian cleavage factor I (CF Im) is an essential factor that is required for the first step in pre-mRNA 3′ end processing. Here, we characterize CF Im68 subnuclear distribution and mobility. Fluorescence microscopy reveals that in addition to paraspeckles CF Im68 accumulates in structures that partially overlap with nuclear speckles. Analysis of synchronized cells shows that CF Im68 distribution in speckles and paraspeckles varies during the cell cycle. At an ultrastructural level, CF Im68 is associated with perichromatin fibrils, the sites of active transcription, and concentrates in interchromatin granules-associated zones. We show that CFIm68 colocalizes with bromouridine, RNA polymerase II, and the splicing factor SC35. On inhibition of transcription, endogenous CF Im68 no longer associates with perichromatin fibrils, but it can still be detected in interchromatin granules-associated zones. These observations support the idea that not only splicing but also 3′ end processing occurs cotranscriptionally. Finally, fluorescence recovery after photobleaching analysis reveals that the CF Im68 fraction associated with paraspeckles moves at a rate similar to the more dispersed molecules in the nucleoplasm, demonstrating the dynamic nature of this compartment. These findings suggest that paraspeckles are a functional compartment involved in RNA metabolism in the cell nucleus.
INTRODUCTION
Most functional mRNAs of eukaryotic genes are generated from their primary transcripts (pre-mRNAs) through RNA splicing and 3′ end polyadenylation. Removal of introns occurs in the spliceosome, a complex composed of five small ribonucleoprotein particles (U1, U2, U4/U6, and U5 small nuclear ribonucleoprotein particles [snRNPs]) and many non-snRNPs splicing factors, including members of the arginine-serine (SR) family of proteins. The mature 3′ ends of mRNAs are generated by endonucleolytic cleavage of the pre-mRNA followed by polyadenylation of the upstream cleavage product. Biochemical studies have identified six factors required for efficient processing in vitro: the cleavage and polyadenylation specificity factor (CPSF), the cleavage stimulation factor (CstF), and two cleavage factors, mammalian cleavage factor Im [CF Im] and CF IIm, are necessary for the cleavage reaction. Polyadenylation requires in addition to CPSF, poly(A) polymerase, and the nuclear poly(A) binding protein 1 (PABPN1, previously called PAB II, for review, see Wahle and Rüegsegger, 1999). Other proteins involved in either transcription, such as the carboxy-terminal domain of RNA polymerase II, or capping (nuclear cap-binding complex) and splicing (U2AF65) have been shown to greatly enhance the efficiency of the first step of the reaction (Flaherty et al., 1997; Hirose and Manley, 1998; Millevoi et al., 2002).
The mammalian cell nucleus is a highly structured and dynamic compartment. Many nuclear factors are localized in morphologically well-defined structural units that include the nucleolus and several “nuclear bodies,” such as the Cajal body (Gall, 2000), and the promyelocytic leukemia body (Zhong et al., 2000). Splicing factors (including SR proteins and snRNPs) were shown by fluorescence microscopy studies to localize to the nucleus in 20–50 highly concentrated, punctuate regions referred to as nuclear speckles, which correspond at the electron microscope level to interchromatin granules clusters (IGCs) and perichromatin fibrils (PFs; for review, see Lamond and Spector, 2003). Studies to characterize the polypeptide sequences responsible for speckle localization have identified two motifs that can target proteins to this compartment: the Sm domains of core snRNP proteins and the arginine-serine–rich (RS) domains of SR proteins. SR proteins constitute a family of pre-mRNA splicing factors that play multiple important roles in both constitutive and alternative splicing of pre-mRNAs (for reviews, see Manley and Tacke, 1996; Valcarcel and Green, 1996). The primary structure of SR proteins is composed of one or two N-terminal RNP-type RNA recognition motif (RRM), and a C terminus that consists largely of repeating arginine-serine dipeptides (RS domain).
The 3′ end processing factor CF Im is a heterodimer composed of a small subunit of 25 kDa and one of two larger subunits of 59 and 68 kDa (Rüegsegger et al., 1996). Both larger subunits seem to be highly related in amino acid sequence and have a domain organization that is reminiscent of SR proteins: an N-terminal RNA-binding domain is followed by a proline-rich region, and by a C-terminal, degenerated RS domain in which the RS repeat itself contains RD/E dipeptides. The RD/E dipeptide motif is similar to a phosphorylated RS domain, because the serine residue in the RS repeat is replaced with a negatively charged aspartic acid or glutamic acid residue. Consistent with this observation, the CF Im 68-kDa subunit was identified as a component of isolated IGCs (Saitoh et al., 2004).
We previously reported that all three CF Im subunits (25, 59, and 68 kDa) are nuclear at steady state. Specific antibodies detect the proteins in the nucleoplasm and in discrete foci (Dettwiler et al., 2004). These foci correspond to paraspeckles, a recently described nuclear compartment (Fox et al., 2002). Paraspeckles contain at least three RNA binding proteins that all interact dynamically with the nucleolus in a transcription-dependent manner. In the present study, we have examined the subnuclear distribution of the 68-kDa subunit of CF Im. We have observed that in addition to paraspeckles, CF Im68 can also concentrate in enlarged, speckle-like structures that partially colocalize with nuclear speckles. Using domain deletion mutants fused to the green fluorescent protein (GFP), we have identified sequences that are important for CF Im68 subnuclear localization in speckles and paraspeckles. Electron microscopy demonstrates that CF Im68 is absent from IGCs but that it can be found associated with PFs and interchromatin granules-associated zones (IGAZs; Puvion-Dutilleul et al., 1995). However, upon treatment with 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB), endogenous CF Im68 no longer associates with PFs but it can still be detected in IGAZs. Finally, we have examined the movement of GFP-CF Im68 by using fluorescence recovery after photobleaching (FRAP). We find that the CF Im68 moves at rates up to 100 times slower than free diffusion. Moreover, CF Im68 mobility in and out of paraspeckles is virtually identical to its mobility in the nucleoplasm. These observations imply that CF Im68 is involved in frequent but transient interactions with other nuclear components.
MATERIALS AND METHODS
Cell Culture and Transfection
HeLa cells were cultured in Dulbecco's modified essential medium (DMEM) to 80% confluence and then transfected with 3 μg of either pEGFP-CF Im68, pEGFP-68RRM/RS, or pEGFP-68RS (Dettwiler et al., 2004) by using Effectene transfection reagent (QIAGEN, Valencia, CA) according to the manufacturer's instructions.
At 20–24 h posttransfection, the cells were fixed for 10 min in 4% (wt/vol) paraformaldehyde in 1× phosphate-buffered saline (PBS) at room temperature, mounted on to glass slides, and imaged under microscopes as described below. For all constructs, these cells were transient transfections. Thus, there was variation in the expression levels of the fusion proteins, resulting in variation in the number and intensity of CF Im foci observed, with overexpressing cells showing numerous (>20) very bright nuclear foci and with low expressors resembling the anti-CF Im68/59 antiserum stain (Dettwiler et al., 2004). Actinomycin D treatment (5 μg/ml; Sigma-Aldrich, St. Louis, MO) was carried for 2 h followed by fixation and fluorescence microscopy.
Cell Synchronization and Cell Cycle Analysis
HeLa cells were grown on coverslips in 60-mm Petri dishes and treated according to different synchronization protocols. Serum starvation was carried out for 48 h. Synchronization at the G1/S transition was performed with a double thymidine block protocol (first 19 h of incubation with 2 mM thymidine, an interval of normal medium incubation for 9 h, and second incubation with 2 mM thymidine for 16 h). To enrich for cells in S phase, cells were released from the thymidine block and incubated for 5 h before fixation. G2/M arrest was obtained with a nocodazole protocol (0.3 μM nocodazole incubation for 20 h). Mitotic cells were then removed by tapping on the cell culture dish. After synchronization, coverslips were removed, and the remaining cells were trypsinized, stained with propidium iodide, and analyzed by flow cytometry. Flow cytometric analysis was performed with FACSCalibur (BD Biosciences, San Jose, CA) and with the ModFit LT 3.0 software (Verity Software House, Topsham, ME).
To determine the most frequent subnuclear localization pattern at each cell cycle stage, sequential section of 35–45 cells that had been double labeled for CFIm68 and either SC35 or PSF was recorded on a DM IRE2 confocal microscope (Leica, Wetzlar, Germany).
Preparation of Cell Lysates and Immunoblotting
Confluent 6-cm dishes of cells HeLa were washed with 1× PBS and lysed using radioimmunoprecipitation assay buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.3% Triton, and protease inhibitor cocktail [Roche Diagnostics, Mannheim, Germany]). Lysates were fractionated on a 10% SDS-PAGE. Proteins were subsequently transferred onto nitrocellulose membrane (Whatman Schleicher and Schuell, Dassel, Germany). The membranes were incubated with rabbit anti-CF Im59/68 antiserum (Rüegsegger et al., 1998) or mouse anti-GFP antibody (1:1000 dilution; Roche Diagnostics).
Immunofluorescence Microscopy, Cell Imaging, and Quantification
RNA precursor incorporation was carried out by adding 10 mM bromouridine or 10 mM iodouridine or 2 mM fluorouridine to the medium for 10 min at 37°C.
Cells were washed in 1×PBS and fixed (as described above) at room temperature. Permeabilization was performed with 0.2% Triton X-100 and 0.2% bovine serum albumin (BSA) in 1× PBS for 10 min at room temperature. Immunofluorescence staining was carried out according to standard techniques. Antibodies used were anti-CF Im68 rabbit polyclonal (1:500; Rüegsegger et al., 1998), anti-PSP1 rabbit peptide antibody (dilution 1:500), anti-PSF mouse monoclonal antibody (mAb) (dilution 1:500; Sigma-Aldrich), anti-fibrillarin mouse monoclonal 72b9 (dilution 1:50; Reimer et al., 1987), anti-SC35 mouse monoclonal (dilution 1:1000; Sigma-Aldrich), anti-SR proteins mouse monoclonal 16H3 (dilution 1:250; Zymed Laboratories, South San Francisco, CA), anti-p80 coilin monoclonal 5P10 (dilution 1:50), mouse monoclonal anti-CPSF100 (J1-27, Jenny et al., 1994), chicken anti-CstF64 (1:500), and fluorescein isothiocyanate (FITC)- and tetramethylrhodamine B isothiocyanate-conjugated secondary antibodies (FITC and Cy3, 1:500; Jackson ImmunoResearch, West Grove, PA). Before mounting on slides, coverslips were sometimes soaked in 1 μg/ml 4′,6-diamidino-2-phenylindole (in 1× PBS for 10 min) to stain DNA and/or followed by soaking in pyronin Y (0.66 mM in distilled H2O for 2 s) to stain RNA. Cells were mounted in either Mowiol/Dabco (for confocal microscopy) or with Vectashield (Vector Laboratories, Burlingame, CA, for the DeltaVision system).
Fluorescence microscopy of fixed cells was carried out either with a Zeiss DeltaVision restoration microscope (Applied Precision, LLC, Issaquah, WA) or with a Leica DM IRE2 confocal microscope equipped with an argon/krypton laser (488 nm) to excite GFP fluorescence and Alexa 594 fluorocrome, and (543 nm) to excite Cy3 fluorescence and a 63×, 1.4 oil HCX Plan-Apochromat objective. For double-labeling experiments, images from the same focal plane were sequentially recorded in different channels and superimposed.
Immunoelectron Microscopy
HeLa cells were grown in DMEM medium in 25-cm2 flasks, with the addition of 10% fetal calf serum, 2 mM glutamine, and 100 units/ml streptomycin and penicillin. In some cases, cells were treated with 20 μg/ml DRB (Sigma-Aldrich) for 5 h; other cells were incubated in DMEM containing 1 mM Br-uridine (Sigma-Aldrich) for 15 min and then processed as described below.
For electron microscope immunocytochemistry, the cells were trypsinized, fixed with 4% paraformaldehyde in DMEM at 4°C for 2 h, and rinsed in phosphate buffer, pH 7.2). The specimens were then embedded in 2% low-gelling agarose and placed into 0.5 M NH4Cl solution in buffer for 30 min at 4°C to block free aldehyde groups. Finally, specimens were dehydrated in ethanol at room temperature and embedded in LR White resin.
Ultrathin sections on Formvar-carbon–coated nickel grids were incubated on a drop of normal goat serum (NGS) diluted 1:100 in PBS for 3 min. The following antibodies were used: rabbit polyclonal anti CF Im68, 1:50 (Rüegsegger et al., 1998); mouse monoclonal anti-SC35, 1:30 (Sigma-Aldrich); mouse monoclonal anti-proliferating cell nuclear antigen (PANA), undiluted (ICN Biochemicals, Cleveland, OH); mouse monoclonal anti-RNA polymerase II H5, 1:50 (Research Diagnostic, Concord, MA); mouse monoclonal anti-5-bromo-2′-deoxyuridine (BrdU) B-44 (dilution 1:20; Sigma-Aldrich). The incubation with the antibodies (in PBS, 0.05% Tween, and 0.1% BSA) was performed at 4°C for 17 h. After rinsing with PBS-Tween and PBS, the grids were incubated with NGS as described above. The grids were incubated with goat anti-rabbit IgG (Jackson ImmunoResearch) coupled with 12-nm colloidal gold, diluted 1:20 in PBS. Triple immunolabeling was carried out by incubating thin sections with rabbit anti-CF Im (1:50), chicken anti-CstF (1:200), and mouse anti-BrdU (1:5). The grids were then incubated with goat anti-rabbit (12 nM) goat anti-mouse (6 nM), and donkey anti-chicken (18 nM). All the incubations were carried out for 30 min at room temperature. As a control, some grids were floated on the incubation mixture without the primary antibody, treated as described above, and incubated with the appropriate secondary antibody. All the grids were rinsed with PBS and distilled water and finally stained with the EDTA regressive technique of Bernhard (1969) preferential for RNP-containing nuclear components. Some grids with the sections were floated onto 0.2 M terbium citrate, prepared according to Biggiogera and Fakan (1998), for 1 h at room temperature, and then rapidly rinsed with water and dried. Stained specimens were observed with a Zeiss EM900 electron microscope equipped with a 30-μm objective aperture and operating at 80 kV.
Photobleaching Analysis
Live-cell microscopy was performed on a Leica SP2 AOBS confocal laser-scanning microscope by using the 488-nm line of an argon/krypton laser. All experiments were done at 37°C. A laser power of 2.5% was used in image acquisitions, and 100% was used for photobleaching. Routinely, four single scans were acquired, followed by a single bleach pulse. After the bleach, 20–40 images were taken at intervals of 0.86 s, and any remaining fluorescence in the bleached area was normalized to zero. For quantitative analysis of fluorescence recovery, data were doubly normalized as described by Phair et al. (2004). Determination of mean values, SD, and Student's t test were performed using Microsoft Excel (Microsoft, Redmond, WA).
RESULTS
CF Im68 Concentrates in Speckle-like Structures and in Paraspeckles
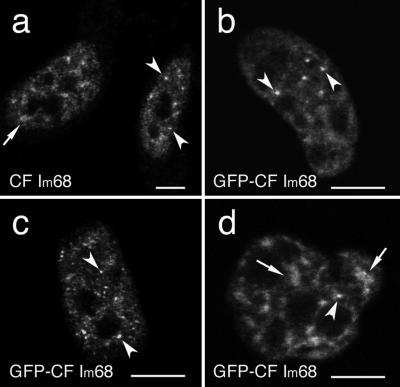
We previously reported that fluorescence microscopy of HeLa cells expressing a GFP-CFIm68 fusion protein shows, in addition to a diffuse nucleoplasmic staining, localization in few discrete foci (Figure 1b) that correspond to paraspeckles (Dettwiler et al., 2004). However, a careful examination of a large number of cells revealed that differences exist between cells. Although in some cells a more punctuate pattern with five to 15 foci/nucleus could be observed, other nuclei showed speckle-like enrichments and only one or two foci/nucleus (Figure 1, compare b and c). The size of the foci ranged between 100 and 300 nm. The speckled enrichments, instead, varied in number and size from cell to cell within the same preparation, ranging from one to eight per nucleus and measuring from 400 nm to 1.5 μm along their widest length. Both structures could also be observed in cells stained with an antiserum that recognizes all CF Im large subunits (Rüegsegger et al., 1998; Figure 1a).
Figure 1.
CF Im 68-kDa subunit is concentrated in speckle-like structures as well as in discrete foci. HeLa cells were either immunolabeled with a polyclonal antibody directed against CF Im68 (a) or transiently transfected with GFP-CF Im68 (b–d). Confocal microscope sections show that CF Im68 is detected in the nucleus, excluding the nucleoli; the protein is diffused in the nucleoplasm with additional concentration in speckled enlargements (broken arrows) and in bright foci (arrowheads). Bar, 10 μm.
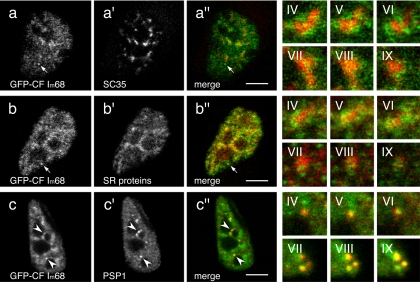
Many, although not all, splicing factors are enriched in nuclear speckles, and the RS domain of SR proteins has been proposed to be a speckle-targeting signal (Lamond and Spector, 2003). Because the CF Im 68-kDa subunit has a C-terminal region enriched in RD, RE, and RS dipeptides that resembles a phosphorylated RS domain, it could be predicted that it may localize to nuclear speckles. Initially, to identify these enlarged enrichments, double-labeling experiments were performed with the anti-CF Im68 antiserum and several different antibodies that identify nuclear speckles (Supplemental Figure S1). However, the polyclonal anti-CF Im68 antiserum recognizes, in addition to the 68-kDa subunit, also the 59- and 72-kDa polypeptides (Rüegsegger et al., 1998). Therefore, to more carefully examine the position of CF Im68-enlarged enrichments relative to nuclear speckles, colabeling experiments were performed with GFP-CF Im68 (Figure 2). For these experiments, we used an antibody to SC35 and an antibody directed against SR proteins that detects SRp75, SRp55, SRp40, and SRp20 but not SRp30a or b (ASF/SF2, SC35). Colocalization experiments of the GFP-CF Im68 fusion protein with the anti-SC35 antibody showed that some of the enlarged enrichments partially coincide with SC35 speckles (Figure 2, a–a″). A similar result was obtained with the anti-SR antibody (Figure 2, b–b″). Analysis of these structures in several different cells revealed that the overlap between GFP-CF Im68 and nuclear speckles was partial and limited to the periphery of the nuclear speckles, as it can be appreciated from the magnifications in Figure 2 (IV–IX, top and middle rows). Perichromatin fibrils have frequently been observed at the periphery of IGC; consequently, localization of CF Im68 at the periphery of nuclear speckles as well as in the nucleoplasm suggests that it may be associated with nascent transcripts. In addition, CF Im68 enrichments were also seen that did not contain SR proteins. In contrast, complete colocalization of GFP-CF Im68 foci with the paraspeckle protein PSP1 was observed in at least 80% of the cases (Figure 2, c–c″). GFP-CF Im68 foci were often found at the edge of nuclear speckles, but they could also be seen in isolation. To determine whether some of the foci could correspond to the previously characterized “cleavage bodies” (Schul et al., 1996), we performed double immunolabeling with CF Im68 antiserum and antibodies that recognize two other essential 3′ end processing factors, CPSF and CstF. However, neither CPSF nor CstF is present in paraspeckles (Supplemental Figure S2).
Figure 2.
Interconnection of CF Im68 speckles and nuclear speckles. Confocal microscope sections of HeLa cells that were transiently transfected with pEGFP-CF Im68, and subsequently immunostained with antibodies against SC35 (a′), SR proteins (b′), or PSP1 (c′). The images of the green fluorescence of pEGFP-CF Im68 (a–c) and the red fluorescence of the antibodies are merged in a″, b″, and c″. Three sequential planes of two different high magnification images (IV–VI, and VII–IX) that were taken from different cells demonstrate the degree of overlap between different compartments. Broken arrows indicate CF Im68 foci. Arrowheads indicate colocalization in paraspeckles. Bar, 10 μm.
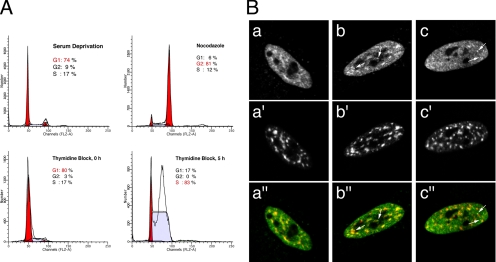
The variability in CF Im68 distribution in foci and speckles suggests that the formation of these structures may be regulated during the cell cycle. To monitor endogenous CF Im68 expression during the cell cycle, we examined cell fractions derived from different synchronization methods: serum deprivation that force cells into quiescence (G0), double thymidine block at G1/S transition, and nocodazole arrest at G2/M. The efficiency of synchronization was monitored by flow cytometry (Figure 3A). Immunofluorescence staining showed a cell cycle-dependent variation in the distribution of CF Im68. Three main phenotypes were observed with variable frequency: diffuse nucleoplasmic staining with speckled enrichments, speckled enrichments with one to three bright foci, diffuse nucleoplasmic staining with several bright foci (Figure 3B, a–c, respectively). Foci-containing cells were enriched at the G1/S transition and in S phase. In contrast, CF Im68 seemed to concentrate in speckles particularly in the nuclei of quiescent cells. In cells arrested at G2/M, CF Im68 localized in speckles as well as in one to three foci. To help with the identification of these structures, cells were double labeled with monoclonal antibodies for SC35 to highlight nuclear speckles, or for PSF, with a protein that was recently shown to colocalize with PSP1 in paraspeckles (Fox et al., 2005). As shown in Figure 3B, the CF Im68-speckled enrichments colocalized with nuclear speckles, whereas the bright foci were often observed adjacent to nuclear speckles (Supplemental Figure S3), and they partially colocalized with the paraspeckles labeled by the anti-PSF antibody (see Figure 7C). These experiments demonstrate that CF Im68 intranuclear distribution is regulated during the cell cycle and suggest that nuclear speckles may represent storage compartments not only for spliceosomal SR proteins but also for the SR-like large subunits of the pre-mRNA 3′ end processing factor CF Im.
Figure 3.
CF Im68 localization is regulated during the cell cycle. HeLa cells were arrested in G0 by serum deprivation, at the G2/M phase using nocodazole, or at the G1/S transition using the double thymidine block procedure (as indicated in A). Cells were then fixed and used for fluorescence-activated cell sorting analysis or double-stained with anti-CF Im68 antiserum as described in Materials and Methods (B, a–c) and anti-SC35 antibody (B, a′–c′). After serum deprivation, CF Im68 concentrated in speckles in 64% of the cells (B, a–a″). In 55% of the cells arrested at G2/M, CF Im68 localized in speckles as well as in one to three foci (B, b–b″). At the G1/S transition and in S-phase CF Im68 was concentrated in foci in ∼60% of the cells (B, c–c″). Quantification of the cellular localization pattern was performed as described in Materials and Methods. The images of the green fluorescence of CF Im68 and the red fluorescence of SC35 are merged in panels a″, b″, and c″. Broken arrows indicate foci.
Figure 7.
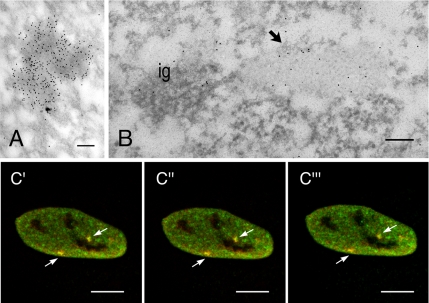
CF Im68 and PSF colocalize in IGAZs and paraspeckles. (A) Immunolabeling for PSF concentrates on dense fibrillar areas. Bar, 0.1 μm. (B) HeLa cells were double labeled with a rabbit antiserum that recognizes CF Im68 and with mouse mAb that recognizes PSF. PSF (small grains) colocalizes with CFIm (large grains) on IGAZ (arrow) nearby a cluster of interchromatin granules (ig). Bar, 0.1 μm. (C) Fluorescence micrographs of three sequential planes sections through HeLa cells that were double labeled for CF Im68 and for PSF. The images of the green fluorescence of CF Im68 and the red fluorescence of PSF are merged in c′–c‴. Broken arrows indicate colocalization in paraspeckles. Bar, 10 μm.
CF Im68 Localization Is Dependent on Ongoing Transcription
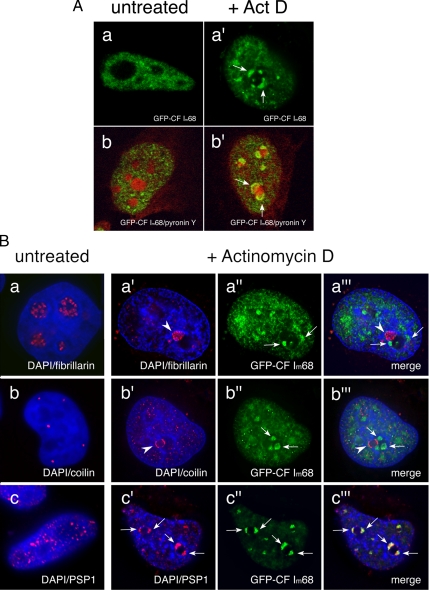
After inhibition of transcription, nuclear speckles become more prominent (Carmo-Fonseca et al., 1992; Spector et al., 1993), whereas paraspeckles' markers, such as PSP1 or PSF, relocalize to perinucleolar caps (Fox et al., 2002; Shav-Tal et al., 2005). We therefore went on to determine the effect of transcription inhibition on CF Im68 localization. HeLa cells, transiently transfected with GFP-CF Im68, were incubated in the presence of 5 μg/ml actinomycin D for 2 h. High actinomycin D concentrations are known to block all nuclear transcription by preventing RNA elongation. As shown in Figure 4A, upon actinomycin D treatment GFP-CF Im68 relocalized, forming very large nucleoplasmic enrichments and distinct “rosettes” (Figure 4A, top row, a′). Staining of cells with pyronin Y that specifically labels RNA demonstrated that the relocalization in cap structures occurred around the nucleoli (Figure 4A, bottom row, b′). The pattern observed for CF Im68 is reminiscent of the relocalization to nucleolar caps of other nuclear proteins, including coilin, and PSP1, after treatment with transcription inhibitors (Raska et al., 1990; Carmo-Fonseca et al., 1992; Andersen et al., 2002). Coilin and PSP1 are known to relocalize to different perinucleolar cap structures (Fox et al., 2002). We therefore performed double-labeling experiments in actinomycin D-treated HeLa cells expressing GFP-CF Im68 with anti-fibrillarin, anti-coilin, or anti-PSP1 antibodies (Figure 4B, top, middle, and bottom, respectively). These experiments showed that only CF Im68 and PSP1 were present in the same perinucleolar cap structures upon inhibition of transcription. This result is consistent with the observation that all other paraspeckle proteins known so far (PSP1, PSP2, PSF, and p54nrb; Fox et al., 2002, 2005; Shav-Tal et al., 2005) relocalize in perinucleolar caps upon RNA polymerase II inhibition where they can be found in the concave caps (Shav-Tal et al., 2005).
Figure 4.
Inhibition of transcription results in the relocalization of CF Im68 to perinucleolar caps. (A) CF Im68 relocalizes in cap structures upon inhibition of transcription. Confocal microscope sections through HeLa cells transiently transfected with GFP-CF Im68 (green) either untreated (a–b) or incubated with 5 μg/ml actinomycin D for 2 h (a′–b′). Cells were stained with pironin Y to show cellular RNA, including nucleoli (red, b and b′). Broken arrows indicate the perinucleolar cap-like structures. (B) CF Im68 colocalizes with PSP1 also after transcription inhibition. Fluorescence micrographs of sections through HeLa cells transiently transfected with GFP-CFIm68 (green) either untreated (a–c) or incubated with 5 μg/ml actinomycin D for 2 h (a′–c‴). Cells were then stained with DAPI to show DNA (blue). Cells were double labeled with antibodies to fibrillarin (red, a and a″), to p80 coilin (red, b and b″), or to PSP1 (red, c and c″). Broken arrows indicate relocalized GFP-CF Im68 at the nucleolar periphery after treatment with actinomycin D; arrowheads indicate relocalized fibrillarin (a and a‴), coilin (b and b‴), and PSP1 (c and c‴), respectively. The sections were obtained using the DeltaVision system, and the images were deconvolved using softWoRx (Applied Precision).
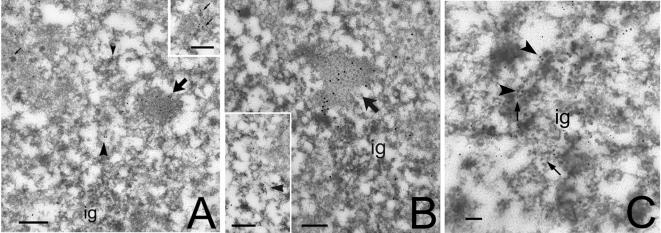
Nuclear speckles correspond to PFs and IGCs at the electron microscopy level. PFs are fibrillar structures measuring 3–5 nm in diameter, and they are found distributed throughout the nucleoplasm and also at the periphery of IGCs (Monneron and Bernhard, 1969). In contrast to IGCs, PFs are labeled rapidly after short pulses with [3H]uridine, suggesting that they represent nascent transcripts (for review, see Fakan, 1994). Recently, CF Im68 was found in purified IGCs (Saitoh et al., 2004). Therefore, to study CF Im68 localization at an ultrastructural level, immunoelectron microscopy was performed on HeLa cells with anti-CF Im68 polyclonal antiserum. As shown in Figure 5A, the gold particles that localized CF Im large subunits accumulated mainly over PFs and perichromatin granules (PGs). Interestingly, PGs were often labeled on their short, emerging tail. Gold particles were also present over compact fibrillar masses that were contiguous with clusters of interchromatin granules (IGs), which in turn were always devoid of labeling. At higher magnifications the dense areas labeled by CF Im that are contiguous with IGCs can be recognized as IGAZs (Visa et al., 1993; Figure 5B). Exclusion of CF Im from IGCs was confirmed by a double-immunolabeling experiment with an anti-PANA antibody (data not shown) that recognizes a marker protein of IGs (Clevenger and Epstein, 1984). This is further confirmed by double labeling with anti-SC35. As shown in Figure 5C, SC35 localized within IGCs as well as on the PFs present at the periphery of the granules and in the nucleoplasm, whereas CF Im could only be found on PFs.
Figure 5.
Ultrastructural localization of CF Im68. HeLa cells were embedded in LR White resin, sectioned, and immunogold labeled with a polyclonal antiserum that recognizes CF Im large subunits (Rüegsegger et al., 1998). Bar, 0.2 μm. (A) At low magnification, a large zone labeled for CF Im (large arrow) is present nearby a cluster of interchromatin granules (ig). The labeling is visible also on perichromatin granules (small arrows) and on PFs (arrowheads). Inset, group of labeled PGs. (B) At higher magnification, the labeled area (large arrow) is recognizable as an IGAZ and is associated with ig. Inset, a single PF is labeled by CFIm. (arrowhead). (C) Double immunolabeling for CFIm (large grains, arrowheads) and SC35 (small grains, broken arrows). Labeling for SC35 is present on the IG cluster, whereas CFIm mostly localizes at the periphery of the IGCs.
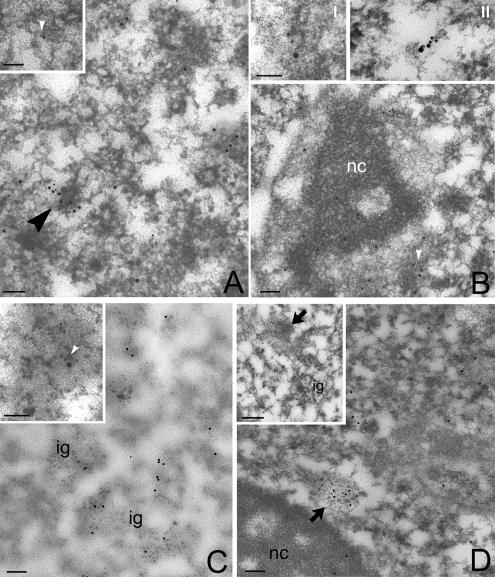
Previous studies have shown that PFs are the in situ form of nascent transcripts and have suggested that they also represent sites of pre-mRNAs processing (Cmarko et al., 1999). The association of CF Im with PFs was tested by colocalization with an anti-RNA polymerase II antibody, which recognizes the active form of the enzyme (Figure 6A). Association of CF Im with nascent transcripts was further verified by labeling newly synthesized RNA with Br-UTP. Both CF Im68 and the 64-kDa subunit of CstF, another essential 3′ end processing factor, colocalized to some extent with BrU-labeled PFs (Figure 6B, inset I and II). Finally, the distribution of CF Im and SC35 was determined in HeLa cells that had been stained with terbium. Terbium staining can be used to specifically visualize RNA-containing nuclear compartments (Biggiogera and Fakan, 1998). As shown in Figure 6C, gold labeling for CF Im colocalized with SC35 over terbium-stained RNA fibrils. In contrast, when RNA polymerase II transcription is inhibited by DRB treatment, immunogold particles were almost exclusively detected on IGAZs (Figure 6D). The weak, residual labeling on PFs disappeared at higher DRB concentrations (data not shown). Together, these results show that both the diffuse nucleoplasmic staining and the overlap of CF Im staining with the periphery of nuclear speckles reflect CF Im association with sites of ongoing transcription and with pre-mRNA molecules. These results confirm and extend previous ultrastructural observations and support the idea that not only splicing (Cmarko et al., 1999; Lamond and Spector, 2003) but also 3′ end processing of pre-mRNAs may occur on PFs.
Figure 6.
CF Im colocalizes with nascent transcripts. (A) RNA polymerase II (small grains) colocalizes with CFIm (large grains) on some PFs (arrowhead). Inset I, at higher magnification, the double labeling is shown on a single, nascent PF. Bar, 0.1 μm. (B) Newly incorporated BrU (small grains) colocalizes with CFIm (large grains) on PFs (arrowhead). Inset, a single, double-labeled PF. Nc, nucleolus. Inset II, a single PF, labeled with anti-BrU antibody (6-nm gold), anti-CF Im68 antiserum (12-nm gold), and anti-CstF antibody (18-nm gold). Bar, 0.1 μm. (C) Newly incorporated BrU (small grains) colocalizes with CFIm (large grains) on PFs (arrowhead). Inset, a single, double-labeled PF. Nc, nucleolus. Bar, 0.1 μm. (D) Immunolabeling for CFIm after selective staining of RNA with terbium. Inset, double immunolabeling for CFIm (large grains) and SC35 (small grains) of terbium-stained RNA. The arrowhead indicates labeling on PFs, whereas interchromatin granules (ig) are unlabeled. Bar, 0.1 μm. (E) HeLa cells treated with DRB. The labeling is decreased on PFs, but still intense on IGAZs (large arrow). Inset, another IGAZ labeled for CFIm after DRB treatment. Ig, interchromatin granules; nc, nucleolus. Bar, 0.1 μm.
In addition, our data suggest that paraspeckles observed by immunofluorescence may correspond to IGAZs. To clarify this point, we examined by immunoelectron microscopy the localization pattern of PSF, a protein that was recently localized to paraspeckles (Fox et al., 2005). This showed gold labeling for PSF concentrated in dense fibrillar domains often associated to IGCs corresponding to IGAZs (Figure 7, A and B). Because double labeling of HeLa cells with anti-CF Im68 and anti-PSF antibodies confirmed that these proteins partially colocalized at the ultrastructural level (Figure 7B), and by confocal laser scanning microscopy (Figure 7C), we conclude that paraspeckles visualized by immunofluorescence microscopy are seen by electron microscopy as IGAZs.
The CF Im68 RNA Recognition Motif Is Required for the Localization in Paraspeckles
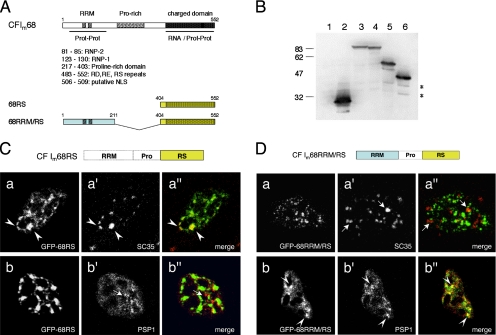
The domain structure of CF Im68 includes an RNA recognition motif of the RNP type at the N terminus (RRM) followed by a proline-rich region and a charged C-terminal domain (“RS-like” domain; Figure 8A). To delineate the sequence requirements for CF Im68 localization to both nuclear speckles and paraspeckles, we analyzed the intranuclear distribution of several domain deletion mutants fused to GFP (Dettwiler et al., 2004). We previously reported that fusion of the C-terminal RS-like domain to the GFP (GFP-68RS, residues 404-552) is sufficient to promote complete nuclear localization of the chimeric protein (Dettwiler et al., 2004). Already at low magnification, the GFP-68RS fusion protein showed, within the nuclear staining, clear speckled enrichments (data not shown). The speckled pattern was much more prominent than that observed with the wild-type protein. To determine whether these structures coincided with nuclear speckles, we performed colocalization experiments with anti-SC35 and anti-PSP1 antibodies (Figure 8). Western blot analysis with an antibody to GFP verified expression in HeLa cells of the full-length protein (GFP-CF Im68), the N-terminal domain (GFP-68RRM, residues 1-211), the C-terminal RS-like domain (GFP-68RS, residues 404-552), and of a fusion of the RRM and the RS domains (GFP-68RRM/RS; Figure 8B). As shown in Figure 8C, the GFP-68RS enrichments were adjacent to PSP1 paraspeckles (bottom row) and completely colocalized with SC35 (Figure 8C, top row). This pattern differs from that observed with the full-length GFP-CF Im68 protein, for which colocalization with SC35 was limited to the periphery of the speckles (Figure 2, top row).
Figure 8.
CF Im68 deletions mutants identify regions important for the localization in speckles and in paraspeckles. (A) Domain structure of CF Im 68-kDa subunit. CF Im68 contains an N-terminal RNA recognition motif of the RNP type (RRM, amino acids [aa] 81-130), a central proline-rich region (Prorich, aa 217-403), and a C-terminal charged domain similar to the RS domain of SR proteins (aa 404-552) with a putative NLS (black box, aa 506-509). (B) Western blot analysis verifies the expression of GFP-fusion constructs in HeLa cells. Lane 1 is the nontransfected control lysate, lane 2 is GFP (expected molecular weight [MW] 27), lane 3 is GFP-CF Im68 (expected MW 86.1), lane 4 is GFP-68RRM/RS (expected MW 67), lane 5 is GFP-68RRM (expected MW 50.4), and lane 6 is GFP-68RS (expected MW 44.4). (C) The RS-like domain is sufficient for localization in speckles. In the left column are GFP images of HeLa cells transiently transfected with the RS fusion construct. The middle column shows fluorescence images of cells stained with antibodies that recognize either SC35 (a′) or PSP1 (b′). The right column shows the overlays of the GFP images (green) and the antibodies (red). Arrowheads indicate areas of colocalization (yellow). Broken arrows indicate paraspeckles. (D) The RRM is necessary for paraspeckles localization. In the left column are GFP images of HeLa cells after transient transfection of the RRM/RS fusion construct. The middle column shows fluorescence images of cells stained with antibodies that recognize either SC 35 (a′) or PSP1 (b′). The right column shows the overlays of the GFP images (green) and the antibodies (red). Arrowheads indicate areas of colocalization (yellow). Broken arrows indicate SC35 speckles.
The GFP-68RRM/RS chimeric protein instead showed a more punctuate pattern. In this case, staining with anti-SC35 showed at best only a partial overlap (Figure 8D, top row), whereas colocalization with PSP1 was complete (Figure 8D, bottom row). To determine whether the RRM domain is sufficient for paraspeckle localization, we wanted to analyze the intranuclear distribution of GFP-68RRM. Because RRM does not contain a nuclear localization signal, a canonical simian virus 40 nuclear localization signal (NLS) was introduced between the GFP and the RRM sequence (GFP-68RRM/NLS). This fusion protein, however, showed a diffuse nuclear staining with concentration in irregularly shaped enrichments that were similar to those observed with GFP alone and did not colocalize with any of the markers (data not shown).
As we reported previously, the C-terminal RS-like domain contains a putative NLS and is sufficient to promote nuclear import of the GFP (Dettwiler et al., 2004). Here, we show that in addition this region is sufficient to direct the protein to nuclear speckles, whereas the N-terminal RRM is needed for paraspeckle localization. Because both the C-terminal region of CF Im68 and its N-terminal RRM are protein interaction domains (Dettwiler et al., 2004), the distribution of the two mutants may reflect protein interactions with components of these subnuclear compartments.
Localization and Mobility of CF Im68 in Living Cells
Inhibition of RNA polymerase II by DRB treatment does not seem to disrupt the association of CF Im large subunits with the interchromatin granules-associated zones. At the fluorescent microscopy level, IGAZs most likely correspond to the paraspeckles, the discrete foci in which GFP-CF Im68 concentrates. Therefore, the question arises whether these structures may be storage compartments from which CF Im68 is recruited to the nascent pre-mRNA molecules that have to be processed.
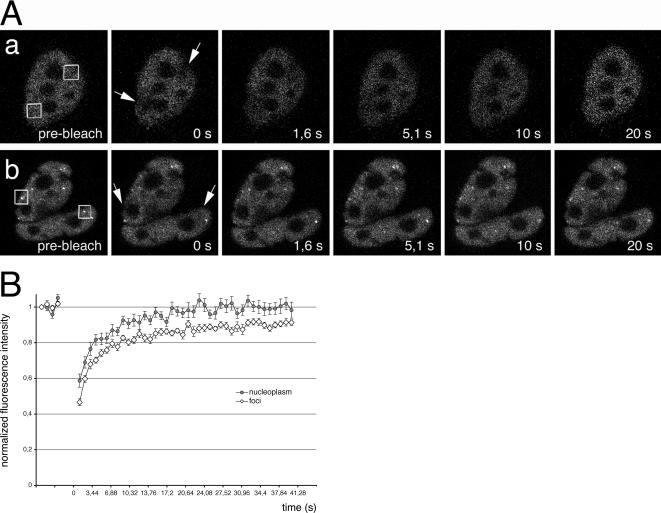
To probe the degree to which CF Im68 is mobile both within the nucleoplasm and in paraspeckles, we carried out FRAP-based experiments in HeLa cells transiently transfected with GFP-CF Im68 (Figure 9A). We analyzed recovery from nucleoplasmic regions, and paraspeckles by bleaching only cells expressing a low level of proteins to avoid overexpression artifacts. Recovery after photobleaching of a nucleoplasmic area was very fast, and the percentage of recovery was high, suggesting the virtual absence of immobile molecules. Recovery of GFP-CF Im68 was almost complete within 20 s, with a half-time of less then 3 s (Figure 9A, top row, and B). GFP-CF Im68 recovery after selective bleaching of a single focus occurred within ∼30 s (Figure 9A, bottom row, and B). An immobile fraction of ∼10% was detected in the paraspeckle, indicating the interaction, or complex formation, of CF Im68 with other components of this subnuclear compartment.
Figure 9.
Recovery of GFP-CF Im68 fluorescence after photobleaching in living cells. (A) Cells expressing GFP-CF Im68 were imaged before or after bleaching of a nucleoplasmic area (top row) or of paraspeckles (bottom row). Images were taken at the indicated times after the bleach pulse. The area of the bleach spot is indicated with a square. a, FRAP of nucleoplasmic regions. b, FRAP of paraspeckles. Bar, 5 μm. (B) Kinetics of recovery after spot bleaching of a nucleoplasmic region (closed circles) and of a paraspeckle (open diamonds). Recovery curves are plotted as normalized fluorescence intensity versus time. Each point is the mean value from eight cells. The error bars indicate the SD.
The mobility of the nucleoplasmic pool of CF Im68 is ∼100-fold lower than that of GFP and slightly but significantly faster than that of paraspeckles-associated CF Im68 (p < 0.01). This may reflect the association of nucleoplasmic CF Im68 with transcripts in perichromatin fibrils and perichromatin granules. These experiments indicate that the CF Im68 fraction that is concentrated in the foci moves at a rate similar to the more dispersed molecules in the nucleoplasm, and they suggest a continuous exchange between the two pools. However, paraspeckles contain a small pool of apparently immobile CF Im68 molecules. Paraspeckles are RNA–protein structures that depend on the RNA for their integrity (Fox et al., 2005). It has been proposed that paraspeckles form as a result of the interaction between paraspeckle proteins and RNA polymerase II transcripts. Therefore, the small immobile pool possibly reflects the engagement of CF Im68 in specific protein–protein interactions that increase its residence time in these structures.
DISCUSSION
In addition to chromatin, the nucleus contains a still increasing number of specific subnuclear compartments (Lamond and Sleeman, 2003). It has recently become clear that some of these compartments are dynamic structures that are capable of moving within the nucleus and of physically interacting with each other. It is also now known that there is a constant flux of molecules into and out of these mobile structures as well as exchange of molecules between them. This is particularly evident for RNA-processing components.
The 68-kDa subunit of the pre-mRNA 3′ end processing factor CF Im, CF Im68, is a modular protein characterized by an N-terminal RRM and a C-terminal–degenerated RS domain in which the RS repeat contains RD/E dipeptides (RS-like domain). We reported previously that the RRM binds CF Im 25-kDa subunit, whereas the RS-like domain interacts with a subset of SR proteins (Dettwiler et al., 2004). In the present study, we show that CF Im68 is found in the nucleoplasm in a diffuse pool, at the periphery of nuclear speckles, and in few discrete foci that correspond to paraspeckles. However, when we examined the localization of the single RS-like domain fused to GFP (GFP-68RS), we observed that it concentrates exclusively in nuclear speckles (and in the nucleoplasm) but not in paraspeckles (Figure 8C), indicating that the C-terminal region is sufficient for the localization in this compartment. Fusion of CF Im68 RRM to the RS-like domain leads to the relocalization of the chimeric protein from nuclear speckles to paraspeckles (Figure 8D). Therefore, the distribution of the full-length protein between nuclear speckles and paraspeckles seems to result from the combination of the protein interactions in which the single domains are engaged.
The RS-like Domain: A Speckle-targeting Signal
Splicing factors show a speckled pattern referred to as the “splicing factor compartment” or “nuclear speckles” (for review, see Lamond and Spector, 2003). The RS motif is the most prominent feature shared by the majority of proteins that have been localized to speckles. For SR proteins that have a single RRM, the RS domain is both necessary and sufficient as speckle-targeting signal (Caceres et al., 1997). Within the nucleus, full-length CF Im68 partially colocalized with SC35, a marker for nuclear speckles. The overlap was generally limited to the periphery of the speckles (Figure 2). At the ultrastructural level, nuclear speckles can be divided into PFs, which contain nascent transcripts and are usually at the periphery of speckles, and IGCs. In contrast to the full-length protein, localization of the isolated RS-like domain was completely overlapping with nuclear speckles, possibly because of the lack of other regions of the protein that are required for CF Im68 function in the maturation of nascent pre-mRNAs (see below).
Recently, 31 proteins with RS motifs, including CF Im68, were identified in isolated IGCs (Saitoh et al., 2004). CF Im68 belongs to a group of four of these proteins that is characterized by RE/RD domains. RE repeats were found in the splicing factor YT521-B that was shown to localize to transcriptionally active sites and was suggested to play a role in grouping genes into higher order structures (Nayler et al., 2000). Thus, proteins with both RS and RD/E motifs may bridge sites of active transcription with IGCs. Our electron microscopy data add support to this hypothesis. We show that CFIm68 is present on nascent PFs that were shown to be the in situ form of nascent transcripts (Cmarko et al., 1999; Trentani et al., 2003) and on the tail of PGs (Figure 5). Moreover, it colocalizes with BrU, RNA polymerase II, the splicing factor SC35, and the 3′ end processing factor CstF (Figure 6). These ultrastructural observations are in agreement with biochemical data indicating that not only splicing but also polyadenylation occurs cotranscriptionally (for review, see Bentley, 2002) and allows the definition of the polarity of the nascent transcript.
Paraspeckles: Storage Compartments or Sites of RNA Posttranscriptional Processing?
Paraspeckles were originally described by immunofluorescence microscopy as the compartment in which the paraspeckle protein 1 (PSP1) accumulates, a novel protein identified in a proteomic study of purified human nucleoli (Fox et al., 2002). In addition to PSP1, four other proteins have been found in paraspeckles: PSP2, p54nrb, PSF, and CF Im68. All these proteins contain RNA-binding motifs, and it was recently shown that PSP1 requires a functional RRM to localize to paraspeckles (Fox et al., 2005). Proteins that bind the RRM might be important in targeting CF Im68 to the paraspeckles and/or in the recruitment of CF Im68 away from nuclear speckles, thus affecting CF Im activity and in turn the level of pre-mRNA 3′ end processing within the cell. Identification of such additional interaction partners will certainly provide better insight into the function of paraspeckles, which is at present ill characterized.
A number of observations suggest that paraspeckles may be involved in RNA metabolism. Paraspeckles are located in the interchromatin nucleoplasmic space, often adjacent to splicing speckles. In this study, we demonstrate that paraspeckles correspond at the electron microscope level to the IGAZ, a nuclear domain consisting of weakly contrasted, densely packed fibrillar material often associated with clusters of IGs (Figure 7; Visa et al., 1993). It was recently shown that some paraspeckles contain S5-phosphorylated RNA polymerase II (Xie et al., 2006). However, only a little of the S2-P RNA polymerase II form or newly made Br-UTP–labeled RNA was found in this compartment, indicating that paraspeckles are not a major site of transcription.
Our FRAP experiments show that the CF Im68 fraction associated with paraspeckles is mobile and is in a constant flux with the nucleoplasmic pool (Figure 9). Therefore, paraspeckles may represent storage compartments from which CFIm68 is recruited to sites of active transcription. Localization of CF Im at the periphery of nuclear speckles and in the nucleoplasm, as well as its association with PFs, may thus reflect its function in cotranscriptional 3′ end cleavage and polyadenylation. Alternatively, localization of CF Im68 in nuclear speckles and in paraspeckles may reflect different roles in the cell nucleus. Interestingly, Spector and colleagues have recently identified a polyadenylated transcript (CTN-RNA) that is diffusely distributed in nucleus and is also localized to paraspeckles in a complex with PSP1 and p54nrb (Prasanth et al., 2005). Under stress conditions, this RNA is posttranscriptionally cleaved at its 3′ untranslated region, releasing a protein-coding mRNA. These observations may point to a role of paraspeckles and of CF Im in the posttranscriptional processing of RNA molecules that are retained in the nucleus. Our immunofluorescence data, however, do not support this hypothesis, because they show that two other essential component of the pre-mRNA 3′end processing complex, CPSF100 and CstF64, do not localize in paraspeckles (Supplemental Figure 3). In contrast, foci enriched in these factors can be found in the vicinity of paraspeckles. One possible explanation for these observations is that paraspeckles and cleavage bodies may represent storage sites for factors, such as CF Im, CPSF, and CstF, from which they are recruited to the nucleoplasm where they participate in mRNA metabolism.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. I. Lamond (Wellcome Trust Biocentre, University of Dundee, United Kingdom) for the generous gift of antibodies against PSP1, coilin, and fibrillarin and C. Milcarek (University of Pittsburgh, Pittsburgh, PA) for anti-CstF antibody. We are grateful to A. Agresti for helpful discussion and to W. Keller and S. Dettwiler for critically reading the manuscript. This study was partially supported by the Progetto Finalizzato del Ministero della Sanità: Studio delle alterazione del processamento degli RNA messaggeri in patologie degenerative del motoneurone, by the Telethon grant GP0020Y01, and by the Cariplo Foundation. S.C. was supported by a short-term European Molecular Biology Organization fellowship.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0846) on January 31, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Almeida F., Saffrich R., Ansorge W., Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- Bernhard W. A new staining procedure for electron microscopical cytology. J. Ultrastruct. Res. 1969;27:250–265. doi: 10.1016/s0022-5320(69)80016-x. [DOI] [PubMed] [Google Scholar]
- Biggiogera M., Fakan S. Fine structural specific visualization of RNA on ultrathin sections. J. Histochem. Cytochem. 1998;46:389–395. doi: 10.1177/002215549804600313. [DOI] [PubMed] [Google Scholar]
- Caceres J. F., Misteli T., Screaton G. R., Spector D. L., Krainer A. R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Pepperkok R., Carvalho M. T., Lamond A. I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger C. V., Epstein A. L. Identification of a nuclear protein component of interchromatin granules using a monoclonal antibody and immunogold electron microscopy. Exp. Cell Res. 1984;151:194–207. doi: 10.1016/0014-4827(84)90368-9. [DOI] [PubMed] [Google Scholar]
- Cmarko D., Verschure P. J., Martin T. E., Dahmus M. E., Krause S., Fu X. D., van Driel R., Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S., Aringhieri C., Cardinale S., Keller W., Barabino S. M. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Flaherty S. M., Fortes P., Izaurralde E., Mattaj I., Gilmartin G. M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A. H., Bond C. S., Lamond A. I. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in a RNA-dependent manner. Mol. Biol. Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A. H., Lam Y. W., Leung A. K., Lyon C. E., Andersen J., Mann M., Lamond A. I. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Gall J. G. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M., Krauss R. D., Chiang L., Valcarcel J., Green M. R., Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Manley J. L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Jenny A., Hauri H. P., Keller W. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol. Cell. Biol. 1994;14:8183–8190. doi: 10.1128/mcb.14.12.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Sleeman J. E. Nuclear substructure and dynamics. Curr. Biol. 2003;13:R825–R828. doi: 10.1016/j.cub.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Lamond A. I., Spector D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Millevoi S., Geraghty F., Idowu B., Tam J. L., Antoniou M., Vagner S. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep. 2002;3:869–874. doi: 10.1093/embo-reports/kvf173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monneron A., Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J. Ultrastruct. Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Nayler O., Hartmann A. M., Stamm S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 2000;150:949–962. doi: 10.1083/jcb.150.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D., Gorski S. A., Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004;375:393–414. doi: 10.1016/s0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- Prasanth K. V., Prasanth S. G., Xuan Z., Hearn S., Freier S. M., Bennett C. F., Zhang M. Q., Spector D. L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F., Besse S., Chan E. K., Tan E. M., Puvion E. p80-coilin: a component of coiled bodies and interchromatin granule-associated zones. J. Cell Sci. 1995;108:1143–1153. doi: 10.1242/jcs.108.3.1143. [DOI] [PubMed] [Google Scholar]
- Raska I., Ochs R. L., Salamin-Michel L. Immunocytochemistry of the cell nucleus. Electron Microsc. Rev. 1990;3:301–353. doi: 10.1016/0892-0354(90)90006-e. [DOI] [PubMed] [Google Scholar]
- Reimer G., Raska I., Tan E. M., Scheer U. Human autoantibodies: probes for nucleolus structure and function. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1987;54:131–143. doi: 10.1007/BF02899205. [DOI] [PubMed] [Google Scholar]
- Rüegsegger U., Beyer K., Keller W. Purification and characterization of human cleavage factor Im involved in 3′ end processing of messenger RNA precursors. J. Biol. Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- Rüegsegger U., Blank D., Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Spahr C. S., Patterson S. D., Bubulya P., Neuwald A. F., Spector D. L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W., Groenhout B., Koberna K., Takagaki Y., Jenny A., Manders E. M., Raska I., van Driel R., de Jong L. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]
- Spector D. L., Landon S., O'Keefe R. T. Organization of RNA polymerase II transcription and pre-mRNA splicing within the mammalian cell nucleus. Biochem. Soc. Trans. 1993;21:918–920. doi: 10.1042/bst0210918. [DOI] [PubMed] [Google Scholar]
- Trentani A., Testillano P. S., Risueno M. C., Biggiogera M. Visualization of transcription sites at the electron microscope. Eur. J. Histochem. 2003;47:195–200. doi: 10.4081/827. [DOI] [PubMed] [Google Scholar]
- Valcarcel J., Green M. R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Visa N., Puvion-Dutilleul F., Bachellerie J. P., Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur. J. Cell Biol. 1993;60:308–321. [PubMed] [Google Scholar]
- Wahle E., Rüegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Xie S. Q., Martin S., Guillot P. V., Bentley D. L., Pombo A. Splicing speckles are not reservoirs of RNA polymerase II, but contain an inactive form, phosphorylated on serine2 residues of the C-terminal domain. Mol. Biol. Cell. 2006;17:1723–1733. doi: 10.1091/mbc.E05-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Salomoni P., Pandolfi P. P. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2000;2:E85–90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.