Abstract
Entry into mitosis is a highly regulated process, promoted by the activated Cyclin B1/Cdk1 complex. Activation of this complex is controlled, in part, by the protein kinase Aurora-A, which is a member of a multigenic serine/threonine kinase family. In normal cells, Aurora-A activity is regulated, at least in part, by degradation through the APC-ubiquitin-proteasome pathway. It has recently been proposed that, in Xenopus, Aurora-A degradation can be inhibited by phosphorylation. It would thus be expected that a phosphatase activity would release this blockade at the end of mitosis. Here, we have shown that the protein phosphatase PP2A and Aurora-A are colocalized at the cell poles during mitosis in human cells and interact within the same complex. Using the PP2A inhibitor okadaic acid and an RNAi approach, we have shown that this interaction is functional within the cell. PP2A/Aurora-A interaction is promoted by an S51D mutation in Aurora-A and inhibited by a phosphomimetic peptide centered around Aurora-A S51, thereby strongly suggesting that PP2A controls Aurora-A degradation by dephosphorylating serine 51 in the A box of the human enzyme.
INTRODUCTION
Protein phosphorylation is a common mechanism involved in the regulation of many cellular processes, by modifying protein affinities, conformations, and/or recruitment to particular cellular localizations. Eukaryotes express many serine/threonine protein kinases that typically have narrow specificities, whereas they have fewer serine/threonine protein phosphatases that typically have broad spectra of action (Zolnierowicz and Bollen, 2000). The diversity in phosphatase functions comes from the ability of the highly conserved catalytic subunits to combine with various regulatory subunits and give rise to a large repertoire of hetero-multimeric holoenzymes (Virshup, 2000).
The protein phosphatase PP2A belongs to the serine/threonine PP2 phosphatase family that also contains PP2B (also named calcineurin) and PP2C (Lechward et al., 2001). PP2A is a hetero-trimeric complex composed of various isoforms of three subunits. The 36-kDa catalytic C subunit associates with the 65-kDa structural A subunit to form the core dimer. This dimer is able to interact with one of the several B-type regulatory proteins (B, B′, and B″), which modulate the holoenzyme activity and substrate specificity (Price and Mumby, 2000; Strack et al., 2002). The A and C subunits are ubiquitously expressed in mammalian cells (Khew-Goodall and Hemmings, 1988; Hemmings et al., 1990; Mayer et al., 1990), whereas the expression of the different B subunit isoforms is tissue specific and can target the trimer to different subcellular compartments (Healy et al., 1991; Mayer et al., 1991; Zolnierowicz et al., 1994; Strack et al., 1999). Numerous functions have been described for the PP2A holoenzyme depending on the B regulatory subunit, including roles in morphogenesis, signal transduction, apoptosis, and cell-cycle progression and control (reviewed by Lechward et al., 2001). Indeed, PP2A is a key enzyme for the initiation and completion of mitosis. For example, the holoenzyme is required both for the G2/M transition (Lee, 1995; Minshull et al., 1996; Karaiskou et al., 1999) and during mitosis itself. In Xenopus eggs extracts, PP2A activity is required for bipolar spindle assembly and the maintenance of short microtubules during metaphase (Tournebize et al., 1997). PP2A has also been proposed to play a role in the metaphase-anaphase transition in both mitosis and meiosis (Chaudhuri et al., 1997; Mailhes et al., 2003). Finally, mass-spectroscopy–based proteomic analyses have revealed that PP2A is present in human interphase centrosomes (Andersen et al., 2003), which are crucial for cell division and cell-cycle progression (Rieder et al., 2001) because they contain key mitotic kinases such as Nek2 (Helps et al., 2000) and Aurora-A (Gopalan et al., 1997; Goepfert and Brinkley, 2000; Blagden and Glover, 2003; Dai et al., 2003).
The mammalian protein kinase Aurora-A is a member of a multigenic serine/threonine kinase family. The founding member of this family, Ipl1p, was identified in yeast through mutations leading to increased chromosome mis-segregation (Francisco and Chan, 1994), and in Drosophila, loss of Aurora function causes the formation of a monopolar spindle due to a centrosome-segregation defect (Glover et al., 1995). Similarly, loss of Aurora-A activity induces a monopolar spindle in Xenopus without blocking mitosis entry (Liu and Ruderman, 2006). In mammals, overexpression of Aurora-A was found in breast cancer (Sen et al., 1997), and its activity is increased in many cancers (breast, ovarian, colon, prostate, neuroblastoma, and cervical cancer cell lines) due to either gene mutation (Kallioniemi et al., 1994; Schlegel et al., 1995) or transcriptional deregulation. Aurora-A localizes to centrosomes, its expression is restricted to G2 and M phases (Gopalan et al., 1997), and its kinase activity is required for mitotic commitment (Marumoto et al., 2002; Hirota et al., 2003). Aurora-A interacts with the phosphatase PP1, which negatively regulates its activity by dephosphorylation (Katayama et al., 2001). Before mitosis entry, the Cdk1/Cyclin B1 complex inactivates PP1, leading indirectly to the activation of Aurora-A. The kinase in turn has a positive feedback on the activity of this complex, which promotes mitosis entry (Hirota et al., 2003).
In normal cells, Aurora-A activity is at least partially controlled by degradation through the APC-ubiquitin-proteasome pathway (Honda et al., 2000). Recently, Littlepage and Ruderman (2002) have proposed a degradation model in Xenopus. According to this model, Aurora-A degradation is inhibited by phosphorylation of serine 53, which would be sustained until the end of mitosis. Thus, Aurora-A degradation would depend on a phosphatase activity distinct from PP1. On the basis of the similarities in the localization and functions of PP2A and Aurora-A during mitosis, we hypothesized that the two proteins may interact at this stage, suggesting a possible role for PP2A in controlling the Aurora-A phosphorylation and/or degradation pathway. We present here data suggesting that the direct dephosphorylation of serine 51 (the human counterpart of serine 53 in Xenopus) by PP2A allows Aurora-A degradation at the end of mitosis.
MATERIALS AND METHODS
Cell Culture, Synchronization, and Tests of Proliferation
Chinese hamster ovary (CHO) cells were grown in Eagle's medium with alpha modification (αMEM) with 10% heat-inactivated bovine serum except where indicated. HeLa cells were grown in DMEM with 10% heat-inactivated bovine serum. For synchronization, exponentially growing cells were blocked in S phase by adding 5 mM thymidine. After 16 h, cells were washed three times in phosphate-buffered saline (PBS) and released in fresh medium for 3 h to allow complete progression through S and G2 phases. Cells were then incubated with 35 ng/ml nocodazole (Sigma-Aldrich, L'isle d'Abeau, France) for 3 h and then washed three times with PBS and reinoculated into fresh medium to allow commitment to mitosis. To compare the abilities of cell lines to proliferate, 2 × 104 cells of each line were seeded in six-well plates and counted every day for 6 d.
Flow Cytometry
Cell cycle analysis was performed using flow cytometry (FACS) of propidium iodide–stained cells. Transfected or control cells (roughly 2 × 106) were incubated in DMEM without serum and supplemented with 1% saponin for 30 min. After a short centrifugation, the cells were resuspended in DMEM containing 1% fetal calf serum, 0.1% saponin, and 50 μg/ml propidium iodide (Sigma-Aldrich). The red fluorescence was analyzed using a FACScan flow cytometer and the CellQuest analysis software (Becton-Dickinson, Le Pont de Claix, France). The percentages of G0/G1 and G2/M cells were calculated relative to total in-cycle cells, and SDs were obtained from three independent experiments.
Immunofluorescence
Synchronized cells were grown on coverslips coated with 0.2 mg/ml poly-d-lysine (Sigma-Aldrich), washed twice in TBS (20 mM Tris, 137 mM NaCl, pH 7.6), and fixed in TBS/4% formaldehyde for 4 min at room temperature. After three washes with TBS, the cells were fixed again by incubation in methanol for 5 min at −20°C and washed three times in TBS. Nonspecific binding was blocked by a 45-min incubation in filtered TBS supplemented with 0.1% Tween 20 and 3% BSA. The coverslips were then incubated overnight at 4°C with mixed primary antibodies diluted in 0.1% Tween-TBS, washed three times in 0.1% Tween-TBS, and incubated for 3 h with mixed secondary antibodies diluted in 0.1% Tween-TBS. Finally, the coverslips were washed three times in 0.1% Tween-TBS, once in TBS, and then mounted in Mowiol 4-88 mounting medium (Roth Sochiel, Lauterbourg, France) containing 10 μg/ml 4′6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). All primary antibodies were used at a dilution of 1:500. The primary mAb IAK1 (anti-Aurora-A) was from BD Biosciences (le Pont de Claix, France; Cat. No. 610939); rabbit polyclonal anti-Aurora-A was from Calbiochem (VWR International, Fontenay-sous-Bois, France; Cat. No. PC742-100); the mouse monoclonal (clone 1D6), rabbit polyclonal anti-PP2A-C were from Upstate (Chemicon International, Chandlers Ford, UK; Cat. Nos. 05-421 and 06-222); and the rabbit polyclonal anti-γ-tubulin was from Santa Cruz Biotechnology (Santa Cruz, CA; Cat. No. SC10732). All secondary antibodies were purchased from Molecular Probes (Eugene, OR) and used at a dilution of 1:1500. They were labeled either with Alexa Fluor 546 or Alexa Fluor 488. Microscopic images were obtained using a Zeiss LSM510 confocal microscope (Thornwood, NY) with a Plan-neofluar 40× ph3 oil objective, NA 1.3.
Vector Constructions, Transfection, and RNA Interference Experiments
A full-length human Aurora-A cDNA in the pEF6/V5-His TOPO TA plasmid (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France) was provided by Dr. F. Hans (Grenoble, France). For site-directed mutagenesis, full-length hAurA was amplified by PCR and cloned into the pEGFP-N1 vector (Invitrogen, Cergy Pontoise, France). Mutations were then generated in the Aurora-A-GFP fusion protein using the QuickChange Site-Directed Mutagenesis Kit (Stratagene Europe, Amsterdam, The Netherlands) with the primers indicated in Table 1.
Table 1.
List of the primers used
| Application | Forward | Reverse |
|---|---|---|
| Mutagenesis | ||
| AuroraA S51D | 5′GGTCTTGTGTCCTGATAATTCTTCCCAGCGC3′ | 5′GCGCTGGGAAGAATTATCAGGACACAAGACC3′ |
| AuroraA R371D | 5′GCATAATCCCAGCCAGGACCCAATGCTCAGAGAAG3′ | 5′CTTCTCTGAGCATTGGGTCCTGGCTGGGATTATGC3′ |
| AuroraA S51A | 5′GGTCTTGTGTCCTGCTAATTCTTCCCAGCGC3′ | 5′GCGCTGGGAAGAATTAGCAGGACACAAGACC3′ |
| RNAi | ||
| hAur A | 5′GATCCCCATTCTTCCCAGCGCGTTCCTTCAAGAGAGGAACGCGCTGGGAAGAATTTTTTGGAAA3′ | 5′AGCTTTTCCAAAAAATTCTTCCCAGCGCGTTCCTCTCTTGAAGGAACGCGCTGGGAAGAATGGG3′ |
| hAur A mismatch | 5′GATCCCCATGCTTCCAAGCGCGTTCCTTCAAGAGAGGAACGCGCTTGGAAGCATTTTTTGGAAA3′ | 5′AGCTTTTCCAAAAAATGCTTCCAAGCGCGTTCCTCTCTTGAAGGAACGCGCTTGGAAGCATGGG3′ |
| hPP2AC | 5′GATCCCCTCTGTGGAGATGTGCATGGTTCAAGAGACCATGCACATCTCCACAGATTTTTGGAAA3′ | 5′AGCTTTTCCAAAAATCTGTGGAGATGTGCATGGTCTCTTGAACCATGCACATCTCCACAGAGGG3′ |
| hPP2AC mismatch | 5′GATCCCCTCTGTGGACATATCCATGGTTCAAGAGACCATGGATATGTCCACAGATTTTTGGAAA3′ | 5′AGCTTTTCCAAAAATCTGTGGACATATCCATGGTCTCTTGAACCATGGATATGTCCACAGAGGG3′ |
| siRNA | ||
| hAur A | 5′AUUCUUCCCAGCGCGUUCC3′ | 5′GGAACGCGCUGGGAAGAAU3′ |
| hAur A mismatch | 5′AUGCUUCCAAGCGCGUUCC3′ | 5′GGAACGCGCUUGGAAGCAU3′ |
| hPP2AC | 5′UCUGUGGAGAUGUGCAUGG3′ | 5′CCAUGCACAUCUCCACAGA3′ |
| hPP2AC mismatch | 5′UCUGUGGACAUAUCCAUGG3′ | 5′CCAUGGAUAUGUCCACAGA3′ |
The interfering, mismatched, or mutated sequences are in boldface. The vector hairpin sequences are in italic.
For RNA interference (RNAi) experiments, 64-base pair primers were cloned into the pSUPER 2 vector encoding Zeocin resistance (to allow selection of stable cell lines) as described by the provider (Oligoengine, Seattle, WA). To knock down Aurora-A, we used primers designed to yield a sequence previously described to down-regulate the kinase upon transient transfection of HeLa cells (Hirota et al., 2003), as shown in Table 1 along with the control mismatch primers. Because no PP2A-C RNAi experiments have been described previously in mammalian cells, we designed a sequence likely to knock down both the α and β isoforms of PP2A-C by aligning the PP2A-Cα and PP2A-Cβ coding sequences with the RNAi oligonucleotide sequence previously described for Drosophila. We chose a sequence that matches both PP2A-C isoforms and designed the primers and control mismatch primers described in Table 1.
Electroporations were performed following Protocol T820 provided for the BTX ElectroSquarePorator (Harvard Apparatus SARL, Les Ulis Cedex, France). HeLa cells were harvested using a mixture of trypsin and 0.5 mM EDTA, washed twice in PBS, and resuspended in PBS at 5 × 106 cells/ml. DNA, 20 μg, was added to a 400-μl sample in a BTX Disposable Cuvette Model 640 with a 4-mm gap. Electroporation was performed using square pulses of 150 V and 99-ms duration. Two pulses were performed. Electroporated cells were immediately transferred to a 24-well plate and incubated for 24 h at 37°C in fresh DMEM supplemented with 10% serum. Stably silenced cell lines [AUR(RNAi) and PP2A(RNAi)] and control lines [AUR(Mm.RNAi) and PP2A(Mm.RNAi)] were selected using Zeocin (Invitrogen) at a final concentration of 1 mg/ml. For transient transfections, HeLa cells in 6-well plates were transfected using ExGen 500 transfection reagent (Euromedex, Souffelweyersheim, France) following the manufacturer's instructions.
For transient RNAi experiments, chemically synthesized RNAs were purchased from Eurogentec (Angers, France) in the desalted, preannealed duplex form. The interfering sequences were identical to those used in the pSuper experiments (see above). One day before transfection, cells were plated in six-well plates and grown at 37°C to 50% confluence in DMEM with 10% serum and without antibiotics. Oligofectamine (Invitrogen) was used for transfections following the manufacturer's instructions. Briefly, a transfection mixture (200 μl) containing siRNA (200 pmol) and Oligofectamine (4 μl) in serum-reduced Opti-MEM (Invitrogen) was added to each well. Each well was subsequently filled to 1 ml with serum-free DMEM. Cells were incubated in this transfection mixture for 4 h and then cultured further in 2 ml of DMEM supplemented with 10% serum. Two rounds of transfection were performed before synchronization and Western blot analysis.
Okadaic Acid Treatment
Okadaic acid (Sigma-Aldrich) was dissolved in DMSO to a final concentration of 1 μM and added to synchronized cells to a final concentration of 1 nM for at least 15 min but never longer than 2 h. After incubation, the cells were washed three times in PBS and prepared for Western blot analysis or immunofluorescence.
Western Blot Analyses
Synchronized cells were lysed in the presence of protease inhibitors either in 25 mM HEPES, pH 7.2, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, or in 25 mM HEPES, pH 7.2, 150 mM NaCl, 5 mM MgCl2, 1% SDS, 1% NP40. Protein quantification was performed using the microBCA method (PerBio Science, Brebières, France). Laemmli denaturing sample buffer (25 μl) was added, and a final volume containing 100 μg of protein was heated for 15 min at 90°C. Proteins were loaded on a 10% SDS gel, run for 1 h at 100 V, and transferred for 1 h at 100 V to a PVDF membrane. Membranes were blocked for 45 min in TBS containing 5% fat free dried milk, 0.1% Tween 20 and then incubated overnight at 4°C in TBS/0.1% Tween 20 containing monoclonal IAK1 antibody diluted 1:250, monoclonal AC-40 anti-actin antibody (Sigma Aldrich; Cat. No. A3853) diluted 1:100, or polyclonal or monoclonal anti-PP2A-C antibodies (see above) diluted 1:1000. Signals were detected using an horseradish peroxide–conjugated goat anti-mouse IgG or goat anti-rabbit IgG antibody and the ECL chemiluminescence procedure (Amersham Biosciences, Piscataway, NJ). Western blot quantifications were performed using ImageMaser VDS-CL and the 1D gel-analysis procedure of the ImageQuant software (Amersham Biosciences) and normalized against actin. The standard deviations were obtained from three to five independent experiments.
Immunoprecipitation and Peptide Competition
HeLa cells were lysed in TBS supplemented either with 1% Triton X-100 or with 1% NP40 and 1% SDS. Protein-G–coupled beads were incubated for 2 h with 3% BSA in PBS at 4°C. A volume of total lysate containing 500 μg of protein was precleared by incubation with 100 μl of protein G/BSA beads for 2 h and then with a nonrelevant rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories Europe, Newmarket, United Kingdom; Cat. No. 005-000-003) for 1 h. The mixture was centrifuged for 2 min at 1600 rpm after each step. The cleared supernatant was then incubated for 2 h with either 8 μg of the polyclonal anti-Aurora-A antibody (see above) or 4 μg of the polyclonal anti-PP2A-C antibody. Protein G–coupled beads, 100 μl, blocked with BSA (see above) were then added, and the mixture was rocked gently overnight at 4°C and then centrifuged for 2 min at 1600 rpm. The immunoprecipitated complex was then washed three times in the lysis buffer. For Western blot analysis, samples of the initial total lysate and of the immunoprecipitation supernatant, each containing 100 μg of total protein, were heated at 90°C in Laemmli sample buffer and loaded onto the gel. The nonspecific precipitate and specific immunoprecipitate were each resuspended in Laemmli sample buffer to a final volume of 20 μl, heated to 90°C, and loaded on the gel.
For the peptide-competition experiment, two peptides were used: a scrambled-sequence peptide (H2N-NSQRRDLSVCP-COOH) and a specific hAURA-D51 peptide (H2N-RVLCPDNSSQR-COOH), which mimicked the phosphorylated form of the Ser-51–containing peptide. The HeLa cell lysate was precleared as described above and split into three fractions containing 500 μg of total protein apiece. The first fraction was incubated only with the PP2A-C polyclonal antibody, whereas the second and third fractions were incubated for 6 h at 4°C with 100 μM of either scrambled-sequence peptide or hAURA-D51 peptide, respectively, before immunoprecipitation of PP2A-C as described above.
RESULTS
The Catalytic Subunit PP2A-C Is Localized in Centrosomes during Mitosis
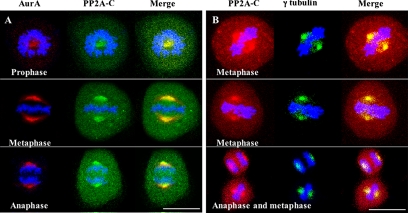
To examine the subcellular localization of the protein phosphatase PP2A, we performed immunofluorescence experiments on synchronized mitotic HeLa cells using confocal microscopy and a mAb directed against the catalytic subunit PP2A-C. The phosphatase was detected ubiquitously throughout the cells but was concentrated at the centrosomes and cell poles (Figure 1, A and B). PP2A-C colocalized approximately with Aurora-A in prophase, metaphase, and anaphase cells (Figure 1A) and with γ-tubulin in metaphase and anaphase cells (Figure 1B), but it occupied a somewhat more restricted area than Aurora A and a somewhat broader area than γ tubulin. The latter results show that PP2A-C is not strictly restricted to centrosomes at the cell poles.
Figure 1.
Colocalization of PP2A-C with Aurora-A at centrosomes during mitosis. (A) Confocal immunofluorescence microscopy of Aurora-A (AurA) using polyclonal anti-Aurora-A antibodies with Alexa Fluor 546–conjugated goat anti-rabbit-IgG (red labeling) and of PP2A catalytic subunit (PP2A-C) using a monoclonal anti-PP2A-C antibody with Alexa Fluor 488–conjugated goat anti-mouse IgG (green labeling) in synchronized mitotic HeLa cells. (B) Same as A except using monoclonal anti-PP2A-C, Alexa Fluor 546–conjugated goat anti-mouse-IgG (red labeling), polyclonal anti-γ-tubulin, and Alexa Fluor 488–conjugated goat anti-rabbit IgG (green labeling). DNA was stained with DAPI. Bars, 20 μm.
PP2A-C Interacts with Aurora-A in Mammalian Cells
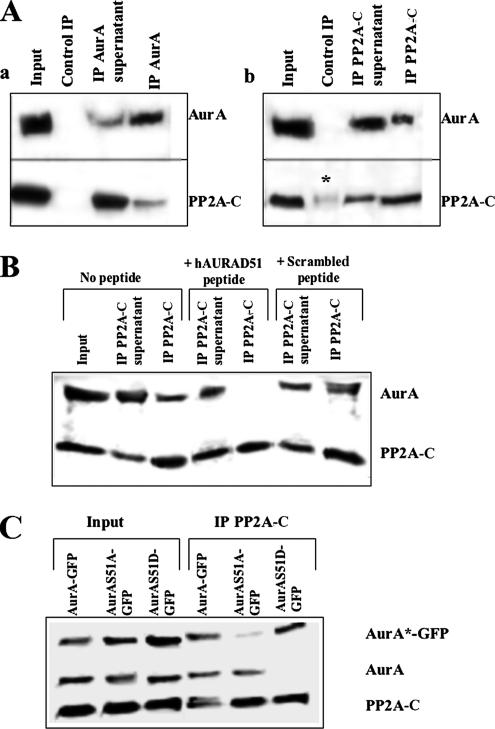
Given the colocalization of PP2A-C with Aurora-A and the requirement of the kinase for dephosphorylation before degradation, we hypothesized that both proteins might belong to the same complex. To address this question, we immunoprecipitated either Aurora-A or PP2A-C from lysates of synchronized mitotic HeLa cells using stringent conditions (1% NP40 and 0.1% SDS) and specific polyclonal antibodies; the precipitates were then analyzed by Western blotting using monoclonal anti-Aurora A or anti-PP2A-C antibodies. When Aurora-A was immunoprecipitated (Figure 2Aa), two proteins with molecular weights of 46 and 35 kDa were observed, corresponding to Aurora-A and PP2A-C. These proteins were also detected in the input and the supernatant but not in the precipitate obtained with an irrelevant immunoglobulin, showing that the coimmunoprecipitation was specific. The smaller amount of PP2A-C than Aurora-A in the immunoprecipitates was consistent with the partial colocalization of the proteins, suggesting that only a limited pool of Aurora-A was associated with PP2A-C at the cell poles. When PP2A-C was immunoprecipitated (Figure 2Ab), both proteins were again present in the precipitate. The coimmunoprecipitation again appeared to be specific, although a faint signal for PP2A-C could be detected when the experiment was carried out with an irrelevant antibody. These reciprocal immunoprecipitation experiments demonstrated that PP2A-C and Aurora-A physically interact, directly or indirectly.
Figure 2.
Coimmunoprecipitation of Aurora-A with PP2A-C and its dependence on Aurora-A phosphoserine 51. (A) Immunoprecipitation (IP) of Aurora-A (a) and of PP2A-C (b) from HeLa cell lysates using polyclonal antibodies and stringent conditions as described in Materials and Methods. A control IP used an irrelevant antibody. Samples were analyzed by Western blotting using monoclonal anti-Aurora-A and monoclonal anti-PP2A-C. Asterisk (*) indicates a low level of nonspecific precipitation of PP2A-C by the control antibody. (B) Interference with coimmunoprecipitation of Aurora-A with PP2A-C by a peptide corresponding to the phosphorylated form of the Aurora-A A box (center lanes). Controls included IP in the absence of added peptide (left lanes) and IP in the presence of the same concentration of a sequence-scrambled peptide (right lanes). Procedures were as in A except for the addition of peptide (see Materials and Methods). (C) Efficient coimmunoprecipitation by anti-PP2A-C polyclonal antibodies of a wild-type Aurora-A-GFP fusion protein and of a fusion protein containing the phosphomimetic S51D mutation but not of a fusion protein containing the nonphosphorylatable S51A mutation. Cells expressing the fusion proteins (see Materials and Methods) were lysed and analyzed as in A.
Littlepage and Ruderman (2002) proposed that Aurora-A degradation depends on dephosphorylation of serine 53 in the A box of the Xenopus protein, suggesting that PP2A-C may interact with a region that encompasses the phospho-serine in this position (serine 51 of the human protein). To test this hypothesis, we performed a competition experiment using an 11-amino acid peptide, hAURAD51, that corresponded to the A box region but with Asp instead of Ser at position 51 in order to mimic the phospho-peptide. A control peptide contained the same amino acids but in a scrambled sequence. Immunoprecipitation of PP2A-C was performed as before but in the presence of these peptides in order to characterize their ability to compete with the endogenous Aurora-A and prevent its association with PP2A-C. As expected, Aurora A was coimmunoprecipitated with PP2A-C in the absence of added peptide or in the presence of the scrambled peptide (Figure 2B). However, peptide hAURAD51 fully inhibited the coimmunoprecipitation (Figure 2B), indicating that the peptide competes efficiently with endogenous Aurora A for interaction with the phosphatase. Thus, the interaction of Aurora A and PP2A-C appears to be direct and requires a region that encompasses the A box.
To determine whether the interaction between PP2A-C and Aurora-A indeed depends on the phosphorylation of serine 51, we generated constructs expressing an Aurora-A-GFP fusion protein and mutant forms of this protein in which S51 was substituted either by aspartic acid (AurAS51D- GFP) or by alanine (AurAS51A-GFP). These constructs were transiently transfected into HeLa cells, and protein expression was monitored by fluorescence microscopy (not shown) and by Western blotting (Figure 2C, left). The Western blots showed that AurA-GFP and AurAS51A-GFP were expressed at comparable levels, that AurAS51D-GFP was expressed at a somewhat higher level, and that the expression of endogenous Aurora-A and PP2A-C appeared to be unaffected. Coimmunoprecipitation experiments using PP2A-C–specific antibody revealed that AurA-GFP and endogenous Aurora- A were both efficiently coimmunoprecipitated, that the S51A mutation weakened the interaction of AurA-GFP with the phosphatase, and, conversely, that the S51D mutation strengthened the interaction sufficiently that the binding of the endogenous Aurora-A to PP2A-C was efficiently competed (Figure 2C, right). Taken together, our data indicate that Aurora-A and PP2A-C interact directly and that phosphorylation of serine 51 of the A box promotes this interaction.
PP2A-C Triggers Aurora-A Degradation
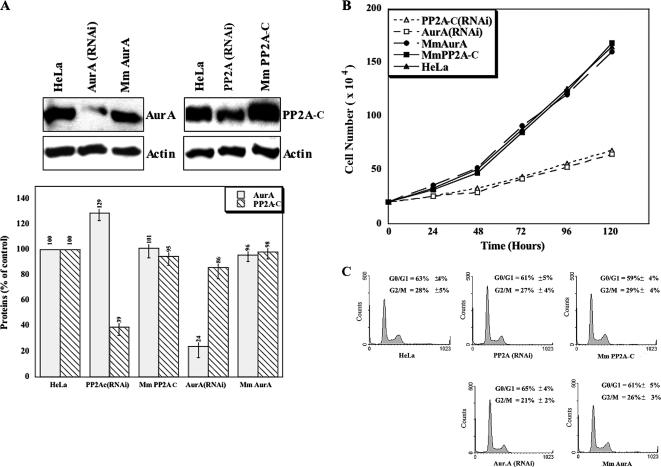
Constructs expressing RNAi sequences directed against Aurora-A or PP2A-C, or control mismatch sequences, were generated as described in Material and Methods, and stable cell lines were selected after transfection. Quantitative Western blot analysis of these cell lines (Figure 3A) revealed that ∼75% of Aurora-A was depleted in AUR(RNAi) cells and ∼60% of PP2A-C was depleted in PP2A(RNAi) cells, whereas the mismatch constructs had no effect, confirming the specificity of the RNAi. Consistent with the hypothesized role of PP2A in Aurora-A degradation, expression of the kinase was increased by ∼30% upon depletion of PP2A-C (Figure 3A). Interestingly, PP2A-C expression was also slightly diminished upon Aurora-A knockdown. We also performed parallel experiments using transient transfection with siRNA constructs (see Materials and Methods), obtaining identical results (Supplementary Figure S1).
Figure 3.
Stabilization of Aurora-A by PP2A-C knockdown. (A) Lysates of stable cell lines knocked down for either Aurora-A (left three lanes) or PP2A-C (right three lanes) were examined by Western blotting (top panel) using monoclonal anti-Aurora-A and PP2A-C antibodies. Mm, lines expressing mismatch RNAi constructs (see Materials and Methods). Bottom panel, quantification of the Western blots by densitometry using actin as the loading control. (B and C) Cell-proliferation rates (B) and flow cytometry analysis of cell cycle distributions (C) of wild-type HeLa cells and the stably transfected cell lines knocked down for Aurora-A or PP2A-C or expressing the corresponding mismatch RNAi.
The observed changes in protein-expression levels might have been due to cell-proliferation arrest or an accumulation of the cells in a particular phase of the cell cycle. For instance, blockage in G2/M would presumably result in high Aurora-A expression, whereas blockage in G1 would have the opposite effect. To address this question, we carried out cell-proliferation assays on the various cell lines (Figure 3B). We observed that the population doubling times were ∼48 h for HeLa cells and the two mismatch-construct lines and ∼96 h for the AUR(RNAi) and PP2A(RNAi) lines. Thus, cell proliferation is dramatically slowed by knockdown of these enzymes. However, flow cytometry analyses revealed similar cell cycle distributions (∼30% G2/M cells and ∼60% G0/G1 cells) in each cell population (Figure 3C). Thus, the slower proliferation observed upon PP2A-C or Aurora-A knockdown appears to be due to a general cell cycle delay rather than to an inability of the cells to progress through a particular phase of the cell cycle, in good agreement with other recent data (Liu and Ruderman, 2006). Thus, the increased level of Aurora-A in PP2A(RNAi) cells probably results from impaired degradation due to the knockdown of phosphatase activity.
Aurora-A/PP2A-C Interaction Is Required for Localization of Both Enzymes to the Cell Poles
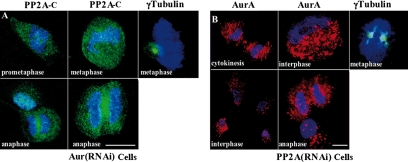
To gain insight into the functional significance of the interaction between Aurora-A and PP2A-C, we analyzed the subcellular localizations of these proteins in AUR(RNAi) and PP2A(RNAi) cells. On Aurora-A knockdown, PP2A-C no longer accumulated at the cell poles during prometaphase, metaphase, and anaphase (Figure 4A). Centrosomes were still present and could be observed by γ-tubulin staining (Figure 4A), but they formed monopolar spindles typical of Aurora-A inhibition or depletion, as observed also by others (Liu and Ruderman, 2006). Thus, the recruitment of PP2A-C to the cell poles of normal mitotic HeLa cells depends on Aurora-A. Reciprocally, Aurora-A was not only stabilized as expected in PP2A(RNAi) cells, but it was also delocalized throughout the cells (Figure 4B), although centrosomes were present at both cell poles, as observed by γ-tubulin staining.
Figure 4.
Interdependent localization of Aurora-A and PP2A-C to centrosomes. Immunofluorescence was performed as in Figure 1 on the stable Aurora-A knockdown line (A) and the stable PP2A-C knockdown line (B) using monoclonal anti-PP2A-C (A) or monoclonal anti-Aurora-A (B) and goat anti-mouse IgG conjugated to Alexa Fluor 488 or Alexa Fluor 546. In separate experiments on each cell line, γ-tubulin was stained using a polyclonal antibody and secondary antibody conjugated to Alexa Fluor 488. Bars, 20 μm.
Aurora-A Stabilization Involves a Decrease in PP2A Enzymatic Activity
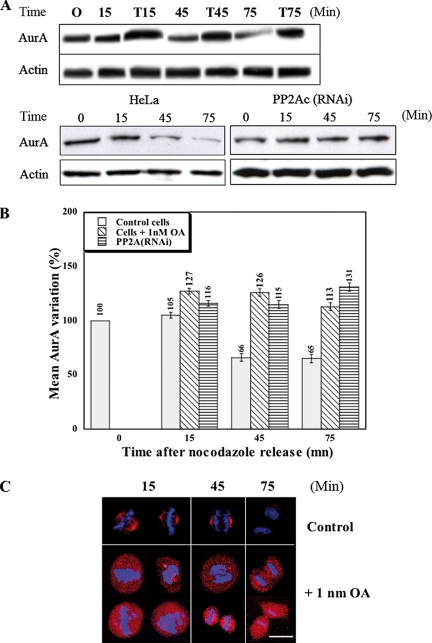
The data presented above suggest that the enzymatic activity of PP2A is required for the degradation of Aurora-A. To examine this possibility more directly, we asked if Aurora-A degradation was inhibited by the serine/threonine-phosphatase inhibitor okadaic acid (OA), which has been shown to preferentially inhibit PP2A activity at low concentrations (Bialojan and Takai, 1988). In these experiments, synchronized cells were treated with 1 nM OA after release from nocodazole arrest in order to avoid any blockage of mitosis entry due to inhibition of protein phosphatase 1, and general cytotoxicity effects were avoided by restricting treatments to 75 min. Samples collected at different times after the release from arrest (time 0) were analyzed by quantitative Western blotting (Figure 5A, top, and B). Within 15 min, the total amount of Aurora-A increased by ∼20% in treated cells; it then remained stable from 15 to 75 min, whereas it decreased in control cells. Stable knockdown of PP2A-C by RNAi had a similar effect (Figure 5A, bottom, and B), suggesting strongly that the OA effect was due mainly to inhibition of PP2A.
Figure 5.
Inhibition or depletion of PP2A protects Aurora-A from degradation. (A and B) Cells were synchronized by nocodazole arrest and sampled at intervals after release for analysis of lysates by Western blotting using the monoclonal antibodies IAK1 and AC-40 (see Materials and Methods). (A, top panel) HeLa cells were treated (T lanes) or not (other lanes) with 1 nM OA beginning at the time of release. (A, bottom panel) HeLa cells (left) and stable PP2A-C knockdown cells (right) were compared. (B) Three independent experiments like those of A were quantitated by densitometry. Means and SDs are shown. (C) HeLa cells were synchronized and treated with OA as described for A and then examined at different times after release by immunofluorescence using the monoclonal anti-Aurora-A antibody and Alexa Fluor 546–conjugated secondary antibodies. DNA was stained with DAPI. Bar, 20 μm.
These biochemical data were supported by immunofluorescence studies, where the decrease in Aurora-A levels in control cells and its stability in OA-treated cells were even more obvious (Figure 5C). The immunostaining also indicated that Aurora-A was also delocalized from centrosomes throughout the cells by the OA treatment, as described above for PP2A-C(RNAi) cells. In addition, some chromosome-alignment defects were visible in the OA-treated cells, whereas control cells showed normal metaphase and anaphase alignments (Figure 5C). This latter observation seems consistent with previously described phenotypes observed when the Aurora-A gene was amplified or when the protein was overexpressed in some cancer cells (Bischoff and Plowman, 1999; Anand et al., 2003).
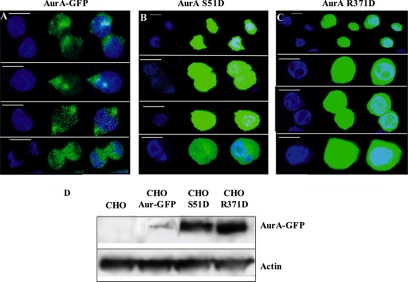
Littlepage and Ruderman (2002) have shown that either substitution of serine 53 of the A box by a residue mimicking phosphorylation or a single point mutation in the degradation box (D box: RxxLxxVxE) is sufficient to stabilize Xenopus Aurora-A in vitro. The former mutation presumably prevents the unmasking of the D box by dephosphorylation of the A Box. As the R residue of the D box is involved in recognition by the APC/Cdh1 complex, its alteration also prevents degradation. It is likely that the mechanism observed in vitro in Xenopus is general and also takes place in mammalian cells in vivo. To address this question, we transiently transfected CHO cells with constructs expressing either wild-type Aurora-A-GFP or one of two mutant versions, AurA(S51D)-GFP and AurA(R371D)-GFP, and compared the susceptibilities of these proteins to degradation. Consistent with the results obtained in the Xenopus studies, both Aurora-A mutants were present at much higher levels than the wild-type protein throughout the cell cycle in the CHO cells, as judged by the GFP fluorescence intensity (Figure 6, A–C). Because the efficiencies of transfection were similar in all cases (∼35%), we could also compare the protein levels by Western blotting, which yielded similar results (Figure 6D).
Figure 6.
Phosphorylation of the A box protects Aurora-A from degradation. (A–C) CHO cells were transiently transfected with constructs expressing the wild-type human Aurora-A-GFP fusion (A), the Aurora-A(S51D)-GFP fusion (B), or the Aurora-A(R371D) fusion (C) and examined for GFP fluorescence. DNA was stained with DAPI. Bars, 20 μm. (D) Western blot analysis of the transfected cells using monoclonal anti-Aurora-A antibody.
DISCUSSION
Aurora-A kinase is a crucial and specific regulator of mitotic events whose protein and activity levels are precisely regulated. It has been reported that Aurora-A kinase activity can be modulated by reversible phosphorylation (for review, see Eyers and Maller, 2003). In particular, Aurora-A is activated at the beginning of mitosis by the LIM protein Ajuba, which promotes Aurora-A autophosphorylation (Hirota et al., 2003). Before this activation stage, Aurora-A is maintained in an inactive, unphosphorylated state by the protein phosphatase PP1 (Katayama et al., 2001). Recent work in Xenopus oocytes also emphasizes that Aurora-A activation depends on kinase rather than phosphatase activities (Maton et al., 2005). In this report, we have shown by immunofluorescence microscopy that Aurora-A and the catalytic subunit of PP2A are colocalized at the cell poles during mitosis and by immunoprecipitation that the two proteins interact within the same complex. These results are consistent with a recent proteomic analysis of interphase human-centrosome components, which reported the presence of PP2A in this organelle (Andersen et al., 2003). Peptide-competition experiments strongly suggest that the interaction between PP2A and Aurora-A is direct and involves at least a small peptide sequence around phosphoserine 51 in the A degradation box. This conclusion is supported by the observation that the Aurora-A S51D point mutation, which mimics phosphorylation, increases PP2A-C affinity for the kinase.
RNAi knockdown of either Aurora-A or PP2A-C indicated that the interaction between these proteins is required to target both proteins to the cell poles. Dephosphorylation of Aurora-A serine 51 by PP2A also appears to be required for the degradation of Aurora-A at the end of mitosis, as shown by the stabilization of Aurora-A by OA treatment at a concentration that preferentially inactivates PP2A, RNAi silencing of the two PP2A-C subunits in HeLa cells, or the S51D mutation. Taken together, our results support the hypothesis that the interaction between Aurora-A and PP2A is physiologically significant, and is involved in the degradation of the kinase at the end of mitosis.
Silencing either Aurora-A or PP2A resulted in a decrease in cell proliferation. Because both Aurora-A and PP2A are involved in mitosis entry, metaphase alignment, and cytokinesis, it is not easy to ascribe these defects specifically to one of these enzymes. However, our results may provide a molecular basis to explain the similar phenotypes described in the literature whenever either Aurora-A or PP2A-C expression is deregulated (Goepfert et al., 2002; Chen et al., 2004; Meraldi et al., 2004). If both proteins are segregated within the same complex, overexpression of one of the partners should result in the delocalization of that protein. PP2A inhibition by either OA treatment or RNAi stabilized but also delocalized the kinase throughout the cell. Thus, phosphorylation of serine 51 within the A box may maintain a structural conformation not only unfavorable for recognition by the proteasome complex but also essential for kinase localization. It is possible that the centrosomal localization of Aurora-A may be required for the degradation process by allowing the interaction of the kinase with some mediator(s) of proteolysis.
Finally, although it is well established that Aurora-A kinase is a crucial regulator of mitosis, the role of PP2A has remained unclear. Our data clearly show that PP2A is a key regulator of Aurora-A degradation and generalize the mechanism suggested by the pioneering studies of Ruderman's group in Xenopus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. J. Lacoste, S. Dimitrov, and W. Herr for helpful comments on the manuscript and Dr. J. Pringle for his careful revision of the English and advice. V.H. was the recipient of a fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur. We are grateful to the Fondation de la Recherche Médicale for supporting the final stage of this work.
Abbreviations used:
- αMEM
Eagle's medium with alpha modification
- DAPI
4′,6-diamidino-2-phenylindole
- FACS
fluorescence-activated cell sorter
- PP2A
protein phosphatase 2A
- RNAi
RNA interference
- siRNA
small interfering RNA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1152) on January 17, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Anand S., Penrhyn-Lowe S., Venkitaraman A. R. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Bialojan C., Takai A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem. J. 1988;256:283–290. doi: 10.1042/bj2560283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff J. R., Plowman G. D. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- Blagden S. P., Glover D. M. Polar expeditions—provisioning the centrosome for mitosis. Nat. Cell Biol. 2003;5:505–511. doi: 10.1038/ncb0603-505. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S. K., Ghosh S., Paweletz N., Schroeter D. Effects of low concentrations of okadaic acid in HeLa cells. Indian J. Exp. Biol. 1997;35:1044–1054. [PubMed] [Google Scholar]
- Chen W., Possemato R., Campbell K. T., Plattner C. A., Pallas D. C., Hahn W. C. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell. 2004;5:127–136. doi: 10.1016/s1535-6108(04)00026-1. [DOI] [PubMed] [Google Scholar]
- Dai W., Huang X., Ruan Q. Polo-like kinases in cell cycle checkpoint control. Front. Biosci. 2003;8:d1128–1133. doi: 10.2741/1129. [DOI] [PubMed] [Google Scholar]
- Eyers P. A., Maller J. L. Regulating the regulators: Aurora A activation and mitosis. Cell Cycle. 2003;2:287–289. [PubMed] [Google Scholar]
- Francisco L., Chan C. S. Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell. Mol. Biol. Res. 1994;40:207–213. [PubMed] [Google Scholar]
- Glover D. M., Leibowitz M. H., McLean D. A., Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Goepfert T. M., Adigun Y. E., Zhong L., Gay J., Medina D., Brinkley W. R. Centrosome amplification and overexpression of aurora A are early events in rat mammary carcinogenesis. Cancer Res. 2002;62:4115–4122. [PubMed] [Google Scholar]
- Goepfert T. M., Brinkley B. R. The centrosome-associated Aurora/Ipl-like kinase family. Curr. Top. Dev. Biol. 2000;49:331–342. doi: 10.1016/s0070-2153(99)49016-7. [DOI] [PubMed] [Google Scholar]
- Gopalan G., Chan C. S., Donovan P. J. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J. Cell Biol. 1997;138:643–656. doi: 10.1083/jcb.138.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy A. M., Zolnierowicz S., Stapleton A. E., Goebl M., DePaoli-Roach A. A., Pringle J. R. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 1991;11:5767–5780. doi: 10.1128/mcb.11.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps N. R., Luo X., Barker H. M., Cohen P. T. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem. J. 2000;349:509–518. doi: 10.1042/0264-6021:3490509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings B. A., Adams-Pearson C., Maurer F., Muller P., Goris J., Merlevede W., Hofsteenge J., Stone S. R. α- and β-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure. Biochemistry. 1990;29:3166–3173. doi: 10.1021/bi00465a002. [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- Honda K., Mihara H., Kato Y., Yamaguchi A., Tanaka H., Yasuda H., Furukawa K., Urano T. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene. 2000;19:2812–2819. doi: 10.1038/sj.onc.1203609. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A., Kallioniemi O. P., Piper J., Tanner M., Stokke T., Chen L., Smith H. S., Pinkel D., Gray J. W., Waldman F. M. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc. Natl. Acad. Sci. USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskou A., Jessus C., Brassac T., Ozon R. Phosphatase 2A and polo kinase, two antagonistic regulators of cdc25 activation and MPF auto-amplification. J. Cell Sci. 1999;112:3747–3756. doi: 10.1242/jcs.112.21.3747. [DOI] [PubMed] [Google Scholar]
- Katayama H., Zhou H., Li Q., Tatsuka M., Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J. Biol. Chem. 2001;276:46219–46224. doi: 10.1074/jbc.M107540200. [DOI] [PubMed] [Google Scholar]
- Khew-Goodall Y., Hemmings B. A. Tissue-specific expression of mRNAs encoding α- and β-catalytic subunits of protein phosphatase 2A. FEBS Lett. 1988;238:265–268. doi: 10.1016/0014-5793(88)80493-9. [DOI] [PubMed] [Google Scholar]
- Lechward K., Awotunde O. S., Swiatek W., Muszynska G. Protein phosphatase 2A: variety of forms and diversity of functions. Acta Biochim. Pol. 2001;48:921–933. [PubMed] [Google Scholar]
- Lee T. H. The role of protein phosphatase type-2A in the Xenopus cell cycle: initiation of the G2/M transition. Semin. Cancer Biol. 1995;6:203–209. doi: 10.1006/scbi.1995.0027. [DOI] [PubMed] [Google Scholar]
- Littlepage L. E., Ruderman J. V. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Ruderman J. V. Aurora A, mitotic entry, and spindle bipolarity. Proc. Natl. Acad. Sci. USA. 2006;103:5811–5816. doi: 10.1073/pnas.0601425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailhes J. B., Hilliard C., Fuseler J. W., London S. N. Okadaic acid, an inhibitor of protein phosphatase 1 and 2A, induces premature separation of sister chromatids during meiosis I and aneuploidy in mouse oocytes in vitro. Chromosome Res. 2003;11:619–631. doi: 10.1023/a:1024909119593. [DOI] [PubMed] [Google Scholar]
- Marumoto T., et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- Maton G., Lorca T., Girault J. A., Ozon R., Jessus C. Differential regulation of Cdc2 and Aurora-A in Xenopus oocytes: a crucial role of phosphatase 2A. J. Cell Sci. 2005;118:2485–2494. doi: 10.1242/jcs.02370. [DOI] [PubMed] [Google Scholar]
- Mayer R. E., Hendrix P., Cron P., Matthies R., Stone S. R., Goris J., Merlevede W., Hofsteenge J., Hemmings B. A. Structure of the 55-kDa regulatory subunit of protein phosphatase 2A: evidence for a neuronal-specific isoform. Biochemistry. 1991;30:3589–3597. doi: 10.1021/bi00229a001. [DOI] [PubMed] [Google Scholar]
- Mayer R. E., Khew-Goodall Y., Stone S. R., Hemmings B. A. Expression and organization of protein phosphatase 2A catalytic subunit genes. Adv. Second Messenger Phosphoprotein Res. 1990;24:236–241. [PubMed] [Google Scholar]
- Meraldi P., Honda R., Nigg E. A. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr. Opin. Genet. Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Minshull J., Straight A., Rudner A. D., Dernburg A. F., Belmont A., Murray A. W. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Price N. E., Mumby M. C. Effects of regulatory subunits on the kinetics of protein phosphatase 2A. Biochemistry. 2000;39:11312–11318. doi: 10.1021/bi0008478. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Faruki S., Khodjakov A. The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 2001;11:413–419. doi: 10.1016/s0962-8924(01)02085-2. [DOI] [PubMed] [Google Scholar]
- Schlegel J., Stumm G., Scherthan H., Bocker T., Zirngibl H., Ruschoff J., Hofstadter F. Comparative genomic in situ hybridization of colon carcinomas with replication error. Cancer Res. 1995;55:6002–6005. [PubMed] [Google Scholar]
- Sen S., Zhou H., White R. A. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- Strack S., Chang D., Zaucha J. A., Colbran R. J., Wadzinski B. E. Cloning and characterization of Bδ, a novel regulatory subunit of protein phosphatase 2A. FEBS Lett. 1999;460:462–466. doi: 10.1016/s0014-5793(99)01377-0. [DOI] [PubMed] [Google Scholar]
- Strack S., Ruediger R., Walter G., Dagda R. K., Barwacz C. A., Cribbs J. T. Protein phosphatase 2A holoenzyme assembly: identification of contacts between B-family regulatory and scaffolding A subunits. J. Biol. Chem. 2002;277:20750–20755. doi: 10.1074/jbc.M202992200. [DOI] [PubMed] [Google Scholar]
- Tournebize R., Andersen S. S., Verde F., Doree M., Karsenti E., Hyman A. A. Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J. 1997;16:5537–5549. doi: 10.1093/emboj/16.18.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup D. M. Protein phosphatase 2A: a panoply of enzymes. Cur. Opin. Cell. Biol. 2000;12:180–185. doi: 10.1016/s0955-0674(99)00074-5. [DOI] [PubMed] [Google Scholar]
- Zolnierowicz S., Bollen M. Protein phosphorylation and protein phosphatases. De Panne, Belgium, September 19–24, 1999. EMBO J. 2000;19:483–488. doi: 10.1093/emboj/19.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnierowicz S., Csortos C., Bondor J., Verin A., Mumby M. C., DePaoli-Roach A. A. Diversity in the regulatory B-subunits of protein phosphatase 2A: identification of a novel isoform highly expressed in brain. Biochemistry. 1994;33:11858–11867. doi: 10.1021/bi00205a023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.