Abstract
HuR is a ligand for nuclear mRNAs containing adenylate-uridylate rich elements in the 3′-untranslated region. Once bound to the mRNA, HuR is recognized by adapter proteins which then facilitate nuclear export of the complex. In the cytosol HuR is thought to function to control stability and translation of its ligand message. In the 3T3-L1 cells HuR is constitutively expressed and localized predominantly to the nucleus in the preadipocytes. However within 30 min of exposure to the differentiation stimulus, the HuR content in the cytosol increases consistent with HuR regulating the availability of relevant mRNAs for translation. Using in vitro RNA gel shifts, we have demonstrated that the C/EBPβ message is a ligand for HuR and that the single binding site is an adenylate-uridylate rich element in the 3′untranslated region.
Keywords: CCAAT enhancer binding protein β, HuR, elav, 3T3-L1, adipocyte, differentiation
Introduction
Adipocyte differentiation is a complex process regulated in large part by the temporally controlled expression and activation of numerous transcription factors [1]. Among these proteins, the C/EBP and PPAR families of transcriptional activators have been identified as critical to initiation of the differentiation process as well as maintenance of the adipocyte phenotype [1]. When growth arrested 3T3-L1 preadipocytes are induced to differentiate, the cells re-enter the cell cycle and undergo mitotic clonal expansion followed by cessation of growth and expression of the adipocyte phenotype [1]. During this process C/EBPβ is expressed and is essential not only for mitotic clonal expansion but the transcriptional activation of PPARγ and C/EBPα [2] and the indispensable nature of this carefully timed expression of C/EBPβ has been demonstrated in several studies [3,4].
Messenger RNA export from the nucleus, mRNA turnover as well as translation initiation are important control points in the post-transcriptional regulation of gene expression. At least in part, control of these processes is exerted through recognition of cis elements in the mRNA by specific binding proteins. One of these binding proteins is HuR, which was first reported to bind to AU-rich elements in the 3′UTRs and is a 32-kDa protein belonging to the Hu/ELAV family (embryonic lethal, abnormal vision) of RNA binding proteins [reviewed in 5]. It is localized predominantly to the nucleus but has been demonstrated to shuttle between the nucleus and cytoplasm [6,7] and suggested to function by binding to nascent mRNAs in the nucleus and protecting them from degradation by actively participating in their nucleo-cytoplasmic transport [6] as well as controlling their stability [8]. Thus, HuR appears to be an important regulatory protein that is involved in the posttranscriptional processing of certain mRNAs.
We have previously demonstrated that the C/EBPβ mRNA is a ligand for HuR and that the formation of the complex may, in part, control the adipocyte differentiation process [9]. The identification of a HuR binding site in the C/EBPβ mRNA was accomplished by inspection and use of the COVELS algorithm [10] which predicted a single HuR binding site between bases 1370 and 1386 of the 3′UTR. This region was confirmed to bind HuR through in vitro RNA gel shift analysis as well as immunoprecipitation of HuR containing messenger ribonucleoprotein particles and RTPCR analysis of the bound message [9]. This region exhibits a high A+U content and would be expected to serve as an adenylate-uridylate rich element (ARE), conferring instability to the message [5]. However, a recent report demonstrated that HuR could function to mediate translocation of the CD38 mRNA from the nucleus to the cytosol through interaction with a structured RNA element that does not contain an ARE and is localized to the coding region of this message [11]. Based on this description of a novel HuR binding site coupled to a translocation function we initiated an examination of the entire C/EBPβ mRNA for non ARE related HuR binding sites. The data presented support the existence of a single site in the C/EBPβ mRNA for HuR binding corresponding to the ARE in the 3′UTR.
Materials and Methods
Materials
Dulbecco's modified Eagle's medium (DMEM) was purchased from Gibco/Invitrogen (Grand Island, New York). Bovine calf serum and fetal calf serum were purchased from Hyclone Laboratories (Logan, Utah). The 3T3-L1 cells used in this work were obtained from Howard Green (Harvard University, Boston, MA). The BCA Protein Assay kit, the NE-PER™ Cell Fractionation kit and HALT™ protease inhibitor mix were from Pierce (Rockford, IL). The MaxiScript T7 kit was from Ambion (Austin, TX). The anti-HuR monoclonal antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and the anti-Armenian-Syrian hamster monoclonal antibody was obtained from BD Pharmingen (San Diego, CA). The cDNA for C/EBPβ was obtained from Stephen Farmer (Boston University, Boston MA). Reagents for molecular biology were purchased from Invitrogen (Carlsbad, CA). All other chemicals were of reagent grade and purchased from Sigma-Aldrich Biochemical (St Louis, MO).
Methods-3T3-L1 cell culture
3T3-L1 preadipocytes were cultured, maintained and differentiated as previously described [12].
Isolation of cytosolic and nuclear fractions
The NE-PER™ Cell Fractionation kit was used to isolate cytosolic and nuclear fractions from the 3T3-L1 cells as per manufacturer’s (Pierce) instructions with minor modifications as previously described [9]. The isolated fractions were stored at −80Co until use.
PCR isolation of regions within the C/EBPβ cDNA
Bases 1 through 314 of the cDNA were isolated using the following primers: Forward 5′-GCCCGTTGCCAGGCGCCGCCTTATAAA-3′ and Reverse 5′-GGCTCCAGGTAGGGGCTGAAGTCGA-3′. Bases 288 through 642: Forward 5′-TCGACTTCAGCCCCTACCTGGA-3′ and Reverse 5′-GACAGGCTGCCGCTGCTG-3′. Bases 625 through 957: Forward 5′-CAGCAGCGGCAGCCTGTC-3′ and Reverse 5′-GCTTGAACAAGTTCCGCAGGTG-3′. Bases 940 through1254: Forward 5′-TGCGGAACTTGTTCAAGCA GC-3′ and Reverse 5′-AGTTACACGTGTGTTGCGTAGTCC-3′. Bases 1179 through 1292: Forward 5′-TCGGGACTTTGATGCAATCC-3′ and Reverse 5′-AACATCAACAACCCCGCAG-3′. Bases 1230 through 1500: Forward 5′-GGACTACGCAACACACGTGTAACT-3′and Reverse 5′-TTCTCGAGCGGATCCTTTGGCTTT-3′. The T7 polymerase binding site was included in double stranded templates used in the preparation of riboprobes for RNA gel shift reactions by including the sequence: 5′-GGATCCTAATACGACTCACTATAGGGAGCT-3′.
RNA gel shift analysis
Radiolabeled riboprobes were prepared using the Ambion Maxi-Script T7 kit as per manufacturer’s instructions and used in RNA gel shift assays as we have previously described [9]. When supershifts were performed either anti-Armenian-Syrian hamster IgG or anti-HuR 3A2 monoclonal antibody (2μl) was added to replicate samples after the binding incubation and the incubation continued for an additional 30 min at room temperature. The anti- hamster IgG antibody was used as a specificity control.
Results and Discussion
We first examined the 5′UTR and coding region of the C/EBPβ clone for the formation of complexes with cytosolic or nuclear proteins. The data presented in Fig. 1A indicates that complex formation with cytosolic derived proteins was not detected using RNA probes corresponding to bases 1 through 314 (lanes 1–3), bases 288–642 (lanes 4–6) or bases 625 through 957 (lanes 7–9). Lanes 10–12 demonstrate our ability to detect protein-RNA probe complex formation using a probe corresponding to our previously described [9] HuR binding site (lanes 10 and 11) and identify the protein component as HuR using the anti-HuR antibody (lane 12). Fig. 1B examines binding using nuclear derived protein. The data displayed in Fig. 1A & B indicate that complex formation does not occur between either cytosolic or nuclear protein and the 5′UTR or coding region of the C/EBPβ mRNA.
Fig. 1.
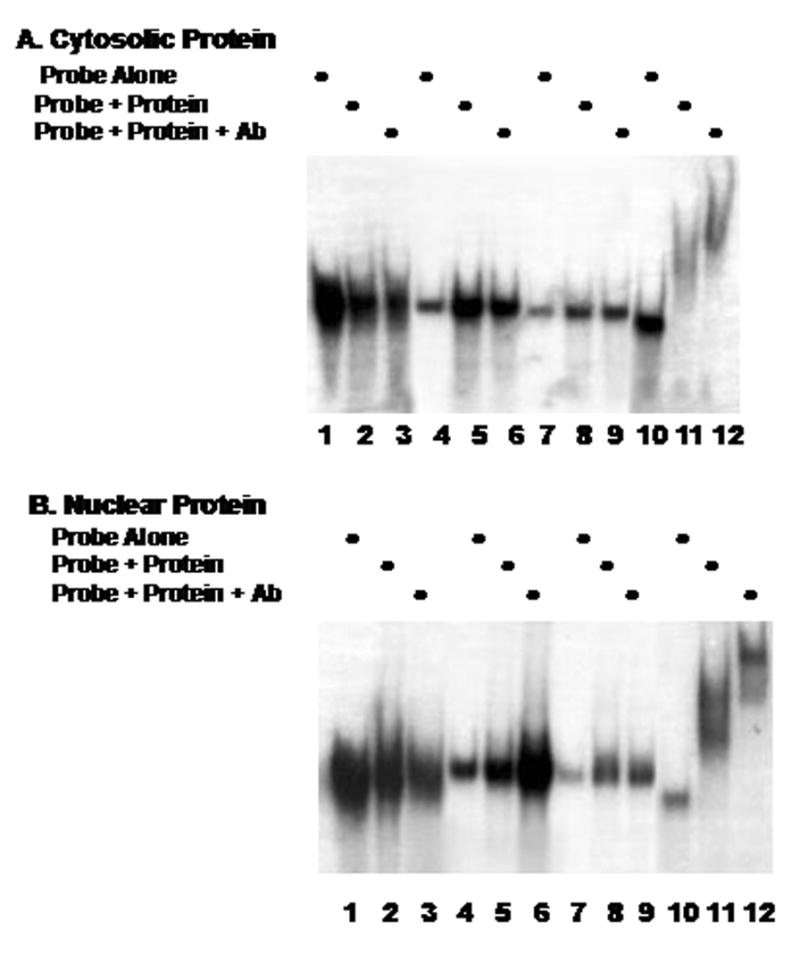
Cytosolic (A) or nuclear (B) protein extracts and radiolabeled probes corresponding to the C/EBPβ 5′-UTR and coding regions (lanes 1–9 as detailed in Methods) were used to perform RNA gel shift and supershift assays (Ab, anti-HuR monoclonal antibody). A radiolabeled probe corresponding to a section of the C/EBPβ 3′-UTR previously demonstrated to bind HuR [1] was included as a positive control, (lanes 10–12).
The 3′UTR was isolated as three sections and the binding of both nuclear and cytosolic protein examined in a series of RNA gel shifts. The data displayed in Fig. 2, indicate that all three regions: bases 940 through 1254, lanes 1–3; bases 1179 through 1292, lanes 4 through 6 and bases 1230 through 1500, lanes 7–9, form complexes with protein from both nuclear and cytosolic compartments as indicated by a significant shift (lanes 2,3,5,6,8,9) relative to the probe alone (lanes 1,4,7). In Fig. 3A, RNA gel shifts were repeated using the same probes and cytosolic protein and supershifts were performed using the anti-HuR 3A2 monoclonal antibody (lanes 3, 6, 9). Only the probe corresponding to bases 1230–1500 generated a supershift (lane 9). This is consistent with HuR binding to the region between 1230–1500 which would correspond to the previously described ARE that serves as the HuR binding site [9]. Fig. 3B repeats the same experiment using nuclear protein and yields results identical to those observed with cytosolic protein. Only the most 3′ region of the message contains a HuR binding site.
Fig. 2.
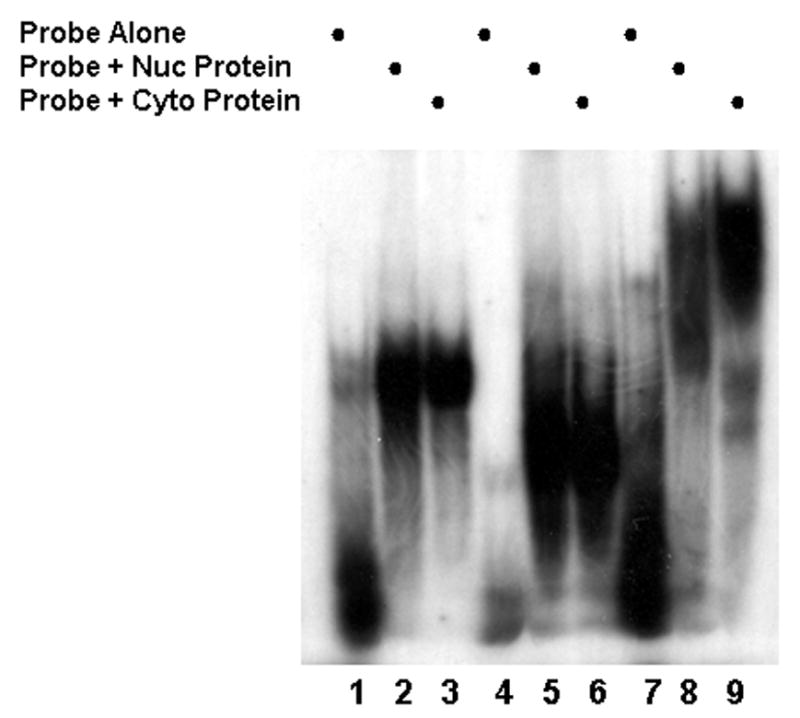
Gel shift assays were performed using radiolabeled probes corresponding to the C/EBPβ 3′-UTR and cytosolic (cyto) or nuclear (nuc) protein extracts.
Fig. 3.
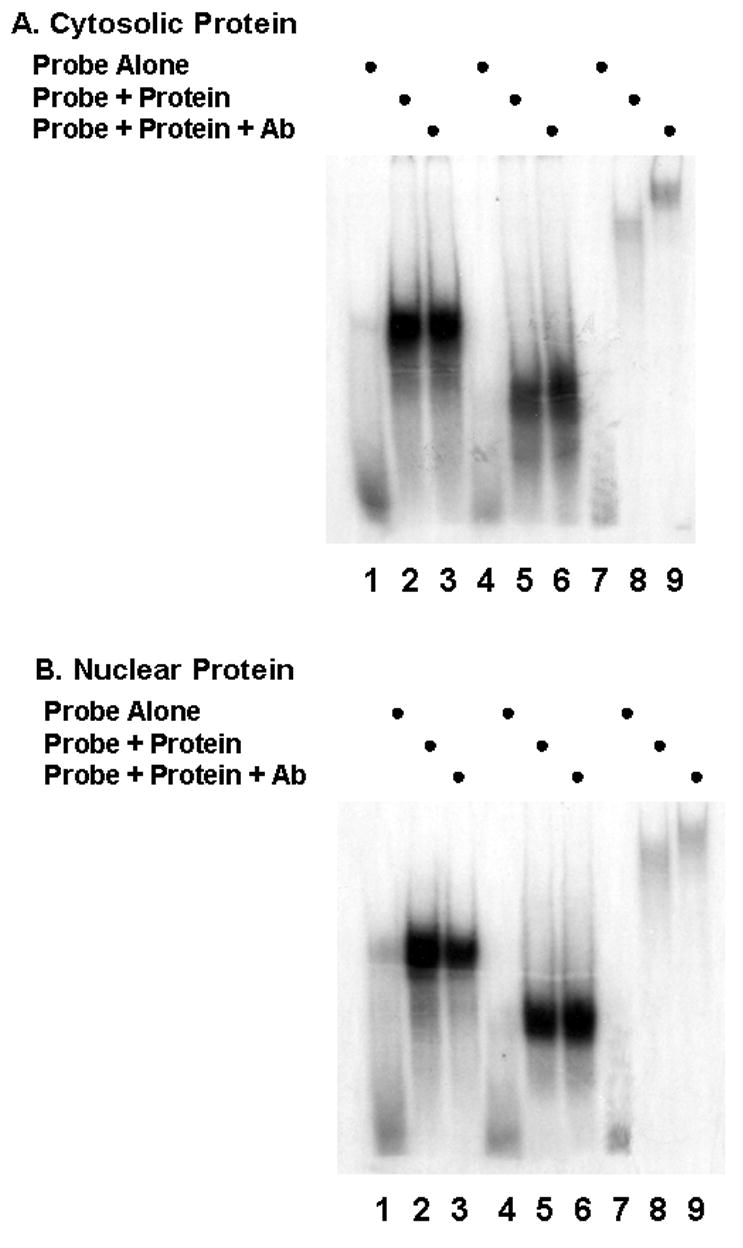
Probes corresponding to the 3′UTR region of C/EBPβ and cytosolic (A) or nuclear (B) protein extracts were used in gel shift assays. Mouse anti-HuR monoclonal antibody (Ab) was included in some experiments.
To further investigate/confirm that the protein complexes formed with the C/EBPβ 3′UTR involved HuR, the previously described binding experiments were repeated and complexes crosslinked using a Strategene UV-Stratalinker, digested with a combination of RNase A and T1 and analyzed for molecular size by SDS-PAGE, The results shown in Fig. 4 demonstrate that the digestion process degrades the probes corresponding to regions 940–1254 and 1179–1292 with no protection of specific regions of the probe by binding proteins. Compare probe alone (lanes 1, 4, 7) with the digest after crosslinking (lanes 2, 3 and 5, 6 and 8,9). Only in the most 3′ fragment (bases 1230–1500) did both nuclear and cytosolic protein protect regions of the probe from RNase degradation (lanes 8, 9 relative to lane 7). The bands observed in lanes 2 and 3 as well as 5 and 6 appear identical to lanes 1 and 4 respectively, suggesting that they represent limit digests of the probe alone and not protected fragments. As expected based on previous work [9] HuR at 36 kDa was present in both nuclear and cytosolic extracts and capable of complex formation. Cytosolic proteins of 66, 76 kDa also bound this region of the C/EBPβ mRNA while complexes with a protein of 89 kDa were weakly observed using both nuclear and cytosolic proteins for the assay. These observations suggest that multiple proteins are involved in complex formation in the distal region of the 3′UTR, particularly once the message is in the cytosol.
Fig. 4.
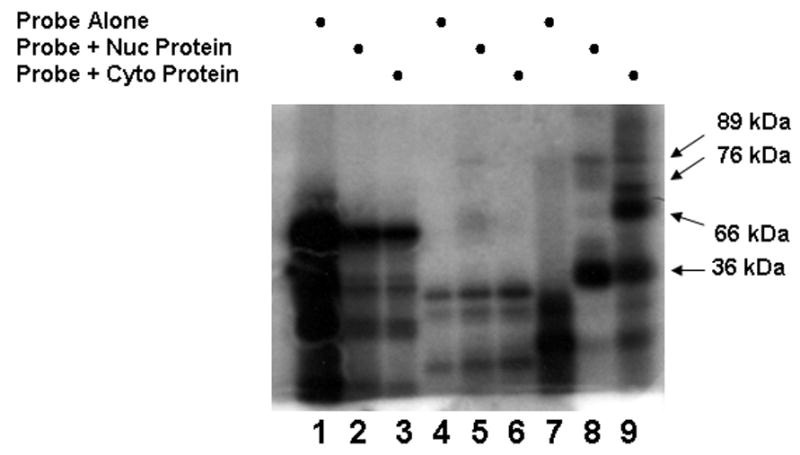
Nuclear (nuc) or cytosolic (cyto) protein extracts and probes for the C/EBPβ 3′UTR region were used in binding experiments, UV-crosslinked and digested with RNase as detailed in Methods. The samples were analyzed for molecular size by SDS-PAGE. Molecular weights of the crosslinked proteins in lanes 7 and 8 were based on the mobility of protein standards.
The data presented in the current study are consistent with: 1) HuR binding to the ARE present in the regions 1230–1500 and 2) that no other HuR binding site exists within the message. Thus, functional regulation mediated by HuR binding to the C/EBPβ mRNA must be mediated through this one binding site.
Acknowledgments
We gratefully acknowledge the computational assistance provided by the laboratory of Dr. Jack Keene, Duke University Medical School for performing the analysis of the C/EBPβ mRNA using the COVELS software package. We further acknowledge support for the performance of these studies provided by: NIH grant DK55169, ADA grant 7-03-RA-76 and a grant from The Brody Brothers Foundation MT7753.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Otto TC, Lane MD. Adipose Development: from stem cell to adipocyte. Crit Rev in Biochem Mol Bio. 2005;40:229–242. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- 2.Zuo Y, Qiang L, Farmer SR. Activation of CCAAT/enhancer binding protein (C/EBP) α expression by C/EBPβ during adipogenesis requires a peroxisome proliferator activated receptor associated repression of HDAC1 a the C/ebpα gene promoter. J Biol Chem. 2006;281:7960–7967. doi: 10.1074/jbc.M510682200. [DOI] [PubMed] [Google Scholar]
- 3.Tang QQ, Otto TC, Lane MD. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci USA. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang JW, Tang QQ, Vinson C, Lane MD. Dominant-negative C/EBP disrupts mitotic clonal expansion and differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 2004;101:43–47. doi: 10.1073/pnas.0307229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherry J, Karschner V, Jones H, Pekala PH. HuR, an RNA binding protein involved in cellular differentiation. In Vivo. 2006;20:17–24. [PubMed] [Google Scholar]
- 6.Myer VE Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO. 1997;16:2130–9. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma WJ Cheng S, Campbell C Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–51. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 8.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gantt K, Cherry J, Tenney T, Karschner V, Pekala PH. An early event in adipogenesis, the nuclear selection of C/EBPβ mRNA by HuR and it’s translocation to the cytosol. J Biol Chem. 2005;280:24768–24774. doi: 10.1074/jbc.M502011200. [DOI] [PubMed] [Google Scholar]
- 10.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prechtel AT, Chemnitz J, Schirmer S, Ehlers C, Langbein-Detsch I, Stulke J, Dabauvalle MD, Kehlenbach RH, Hauber H. Expression of CD83 is regulated by HuR via a novel cis-active coding region RNA element. J Biol Chem. 2006;281:10912–25. doi: 10.1074/jbc.M510306200. [DOI] [PubMed] [Google Scholar]
- 12.Jain RG Andrews LG, McGowan KM Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. MolCellBiol. 1997;7:954–62. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]


