Summary
Correct splice site recognition is critical in pre-mRNA splicing. We find that almost all of a diverse panel of exonic splicing silencer (ESS) elements alter splice site choice when placed between competing sites, consistently inhibiting use of intron-proximal 5′ and 3′ splice sites. Supporting a general role for ESSs in splice site definition, we found that ESSs are both abundant and highly conserved between alternative splice site pairs and that mutation of ESSs located between natural alternative splice site pairs consistently shifted splicing toward the intron-proximal site. Some exonic splicing enhancers (ESEs) promoted use of intron-proximal 5′ splice sites, and tethering of hnRNP A1 and SF2/ASF proteins between competing splice sites mimicked the effects of ESS and ESE elements, respectively. Further, we observed that specific subsets of ESSs had distinct effects on a multifunctional intron retention reporter, and that one of these subsets is likely preferred for regulation of endogenous intron retention events. Together, our findings provide a comprehensive picture of the functions of ESSs in the control of diverse types of splicing decisions.
Introduction
A majority of human genes undergo alternative pre-mRNA splicing, often through alternative inclusion/exclusion of exons (‘exon skipping’) or introns (‘intron retention’) in the processed mRNA. Both of these types of events can have important biological consequences. For example, regulated exon skipping events in the CD44 and Fas receptor genes are involved in control of cell proliferation and apoptosis (Cascino et al., 1996; Cheng and Sharp, 2006). Intron retention is used to regulate the activities of a number of important genes involved in gene expression, including the transcription factor Id3 (Forrest et al., 2004) and the splicing factor 9G8 (Lejeune et al., 2001), and is critical to the life cycles of many viruses (reviewed by Black, 2003).
Another very common type of alternative splicing is the alternative inclusion/exclusion of a portion of an exon, through the use of alternative 5′ splice sites (5′ss) or alternative 3′ splice sites (3′ss) to produce longer and shorter exon forms (Matlin et al., 2005; Sugnet et al., 2004; Yeo et al., 2004). Alternative splice site choice can also be of great consequence. For example, alternative 5′ss selection in the BCL-x gene determines whether the proapoptotic Bcl-x(s) or the anti-apoptotic Bcl-x(L) protein form is produced (Boise et al., 1993), and in Drosophila alternative 3′ss selection in the transformer gene is critical for sex determination (Nagoshi et al., 1988). In human, alternative splice site usage is the most common type of alternative splicing in some tissues (Yeo et al., 2004). Mutations that alter splice site choice are a frequent cause of disease (Baralle and Baralle, 2005; Gabut et al., 2005; Klamt et al., 1998). However, the mechanisms that govern splice site selection generally are not well understood (Black, 2003).
Recently, we used a cell fluorescence-based screening method called FAS-ESS to identify 133 ESS decanucleotide sequences capable of inducing skipping of a constitutive exon in human cells (Wang et al., 2004). A set of 103 hexanucleotides, the ‘FAS-hex3’ set, was significantly enriched in these ESS decamers. These hexamers were shown to commonly possess ESS activity and were found to have substantially reduced frequency in constitutive exons relative to introns and relative to exons subject to exon skipping. The occurrence of distinct peaks in the distribution of these hexamers in introns adjacent to constitutive splice sites suggested a role for ESSs in splice site definition (Wang et al., 2004). Here, we explore this possibility, assessing the effects of the set of FAS-hex3 ESSs, as well as ESEs, on splicing of exons containing either competing 5′ss or competing 3′ss, and determining general rules for ESS activity. We have also explored the roles of ESSs in the regulation of alternative splice site usage and intron retention through a combination of bioinformatic and experimental analyses, identifying additional rules describing ESS activity. The results indicate that many if not all ESSs have potent and predictable effects on several types of alternative splicing that occur commonly in human genes.
Results and Discussion
Our previous analyses of the distribution of the FAS-hex3 ESS hexamers in introns adjacent to constitutive splice sites suggested a role for ESSs in splice site definition (Wang et al., 2004). More detailed analysis of the distribution of these hexamers showed that a peak in ESS density occurs just 3′ of constitutive exons which have a strong downstream ‘decoy’ 5′ss (i.e. a sequence that matches the 5′ss consensus as well as or better than the authentic site), but that this peak is greatly reduced in the absence of decoy 5′ss (Fig. 1A). A peak in ESS density also occurs just 5′ of the branch/3′ss region of exons which have a strong upstream decoy 3′ss, but again no pronounced peak was visible in the absence of a decoy 3′ss (Fig. 1A).
Figure 1. ESSs generally influence splice site choice.
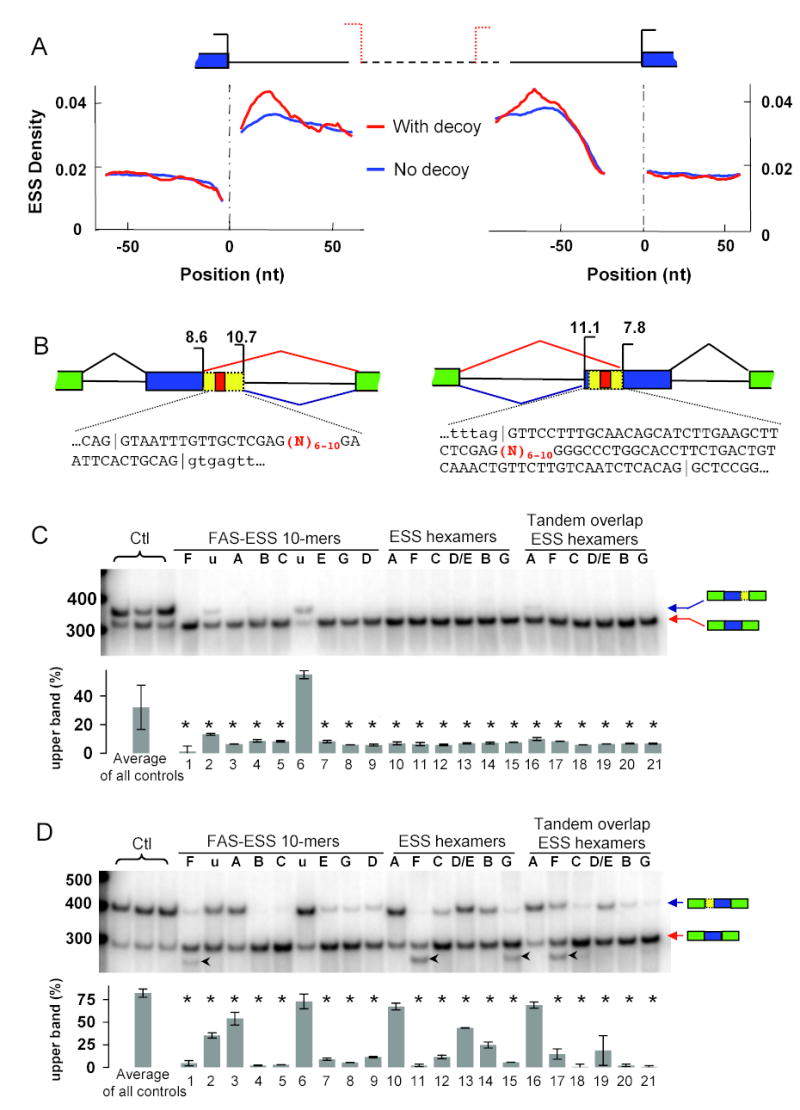
(A) ESS density (FAS-hex3 hexamers as a fraction of total hexamers at each position, smoothed over a window of 10 bases) as a function of distance from the 5′ss and 3′ss of constitutive exons, plotted separately for exons which have (red) or do not have (blue) an adjacent intronic decoy 5′ss or 3′ss (see Supplemental Data). Dataset sizes were, for 3′ss: 11,944 with decoy, 47,048 without decoy; for 5′ss: 7563 with decoy, 59,048 without decoy. Splice sites are shown as left-facing (5′ss) or right-facing (3′ss) brackets, with solid and dotted lines for authentic and decoy splice sites, respectively. (B) Competing 5′ss and 3′ss reporters. Maximum entropy splice site scores (Yeo and Burge, 2004) are indicated above the corresponding splice site brackets (in bits). Sequences between competing splice sites are given (exons in caps, introns lower-case), with splice junctions indicated by ‘|’ and the inserted sequence marked in red. (C) Body-labeled RT-PCR of total RNA from cells transfected with competing 5′ss reporters with primers targeted to flanking exons. All transfections were repeated at least twice. Results shown for ESS sequences representing all 7 FAS-ESS groups (groups A-G + ‘u’ = unclustered; Table S3) and controls (Table S4). Relative usage of the proximal 5′ss is shown below (determined as ratio of intensity of upper band to sum of upper and lower bands). Mean and range of all control sequences is shown at left (seeFig. S1A for all 6 controls); error bars in remaining lanes indicate range of duplicated experiments. All ESS samples except no. 6 (an unclustered decamer) significantly inhibited use of the intron-proximal site (P < 0.02, rank-sum test, indicated by asterisks). Lane 6 appears to be somewhat under-loaded relative to the other lanes based on duplicate transfection data (not shown). (D) RT-PCR results for competing 3′ss reporters (as in panel C); 18 out of 21 ESS inserts significantly inhibited the intron-proximal 3′ss (P < 0.02, indicated by asterisks). For several ESSs that resemble 5′ss, an additional splice form was observed (indicated by arrow head) resulting from use of the inserted sequence as a 5′ss.
ESSs Consistently Inhibit Usage of Intron-Proximal 5′ Splice Sites
These decoy-dependent peaks in ESS density suggested the hypothesis that ESSs function commonly as specificity determinants for splice site recognition by inhibiting the use of intron-proximal decoy 5′ss and 3′ss (i.e. inhibiting downstream decoy 5′ss and upstream decoy 3′ss). To directly test this idea, we constructed splicing reporter systems containing competing 5′ss or 3′ss (Fig. 1B,Table S1). Between competing 5′ss of similar strength, a diverse panel of ESSs was inserted, including: 9 decamers representing the 7 major groups obtained previously by clustering the ESS decamers based on sequence similarity (FAS-ESS groups A through G), and 2 unclustered FAS-ESS decamers; 6 FAS-hex3 hexamers representing consensus sequences of FAS-ESS groups; and 6 decamers designed to contain pairs of overlapping FAS-hex3 hexamers, as well as 6 randomly chosen control decamers (Tables S2, S3). With control sequences inserted between the two 5′ss, ~30% of transcripts were spliced at the intron-proximal 5′ss (Fig. 1C,Fig. S1A,Tables S4, S5). Strikingly, all but one of the 21 ESS elements tested significantly inhibited usage of the intron-proximal 5′ss relative to the distal site (Fig. 1C). These results support the existence of a general rule for splicing regulatory element activity: that sequences capable of inhibiting exon inclusion from an exonic location generally have the ability to inhibit intron-proximal 5′ss when present between competing 5′ss.
Insertion of ESE sequences identified by the RESCUE-ESE method (Fairbrother et al., 2002) between the competing 5′ss gave very different results. The 10 ESEs tested either had no detectable effect on splice site usage or, in 5 of 10 cases, enhanced usage of the intron-proximal 5′ss (Fig. S1B,Tables S4, S5). Thus, when they influenced 5′ss selection, ESEs tended to result in opposite effects of ESSs.
ESSs Consistently Inhibit Usage of Intron-Proximal 3′ Splice Sites
To ask whether 3′ss choice can be similarly controlled by known exonic splicing regulatory elements, a reporter containing competing 3′ss was constructed (Fig. 1B). Insertion of control decamers between the competing 3′ss gave ~80% usage of the intron-proximal 3′ss (Fig. 1D,Fig. S1C). Using the same diverse panel of ESS decamers and hexamers as above, a strong and consistent effect on splice site selection was again observed, with 18 of 21 ESSs tested significantly inhibiting usage of the intron-proximal 3′ss relative to the distal site, often to a relative usage frequency below 25% (Fig. 1D). These data support a generalization of the rule that ESSs inhibit intron-proximal splice sites to include 3′ss as well as 5′ss, with only rare exceptions (discussed in Supplemental Data).
When testing the same set of ten ESEs as above in the competing 3′ss reporter, none had a detectable effect on 3′ss choice (Fig. S1D). Thus, generally speaking, ESSs had more pronounced effects on splice site selection than ESEs in this system.
Conservation and Activity of ESSs Located Between Alternative Splice Sites
The strong and consistent effects on splice site choice of ESSs inserted between competing splice sites suggested that ESSs might commonly play a role in regulation of natural alternative splice site exons. In accord with this idea, a significantly higher density of ESS hexamers was observed in the ‘extension’ regions between alternative 5′ss and between alternative 3′ss (yellow) than in the constant ‘core’ regions of such exons (blue), using large datasets of human alternative 5′ss exons (A5Es) and alternative 3′ss exons (A3Es) (Fig. 2A). To further investigate this phenomenon, we analyzed the degree of conservation of ESS hexamers in the core and extension regions of orthologous human-mouse A5E and A3E exons (1074 and 1318 exon pairs, respectively). Consistent with common function, the number of aligned and conserved ESSs per 100 nt was far higher in extension regions of A5Es and A3Es than in the corresponding core regions (Fig. S2B). Since the precise spacing of ESSs in exons may not be critical for their function (e.g., Han et al., 2005), we also analyzed conservation using a statistic called Conserved Occurrence Rate (COR) which does not require that ESSs in human and mouse extension regions be perfectly aligned in order to be considered conserved (see Methods). Using the COR statistic, we observed that the conservation of FAS-hex3 ESS hexamers in orthologous human/mouse A5E and A3E extension regions was substantially higher than for control sets of hexamers in both classes of exons (P = 4e-12 and P = 5e-4, respectively, Fig. 2B). These results, comparing to control sets of hexamers with identical occurrence counts in A5E and A3E extension regions, indicate the presence of strong selective pressure to conserve occurrence of ESS sequences in these regions.
Figure 2. Role of ESSs in control of alternative splice site choice.
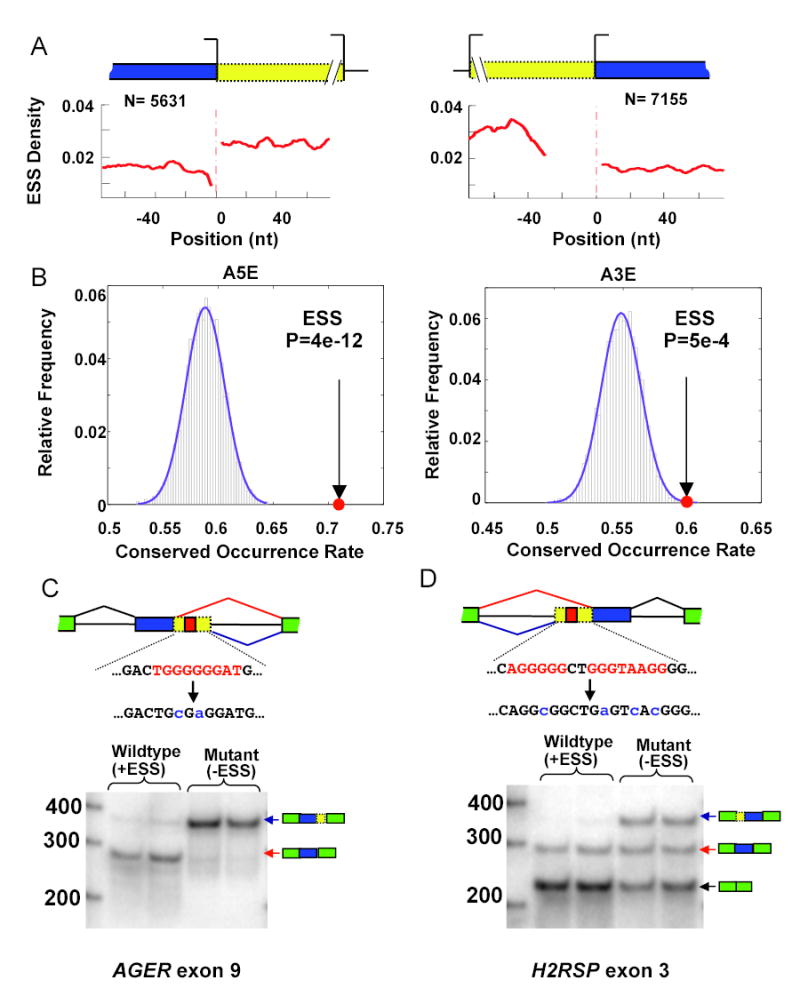
(A) ESS density (as in Fig. 1A) as a function of distance from the intron-distal splice site for human A5Es (left panel) and A3Es (right panel), with the number of exons in each set listed above. (B) Conserved Occurrence Rate (COR, defined in Experimental Procedures) of ESS hexamers in A5E and A3E extension regions (red dot). Histograms show COR values for 5,000 control sets of hexamers of similar abundance in human/mouse extension regions (Supplemental Data). P-values were calculated based on a normal approximation to the distribution of the controls (purple curve). (C) Splicing of A5E minigene based on human AGER gene exon 9 and flanking introns. Partial sequence between alternative 5′ss is shown, with FAS-hex3 hexamers in red, and introduced mutations in blue. Both wild type and mutant constructs were transfected in duplicate (both shown). Splice site usage assayed by body-labeled RT-PCR using primers targeted to flanking exons. (D) Splicing of A3E minigene based on human H2RSP gene exon 3 and flanking introns (same format as panel C).
To directly test the idea that ESS elements are commonly involved in regulation of splice site usage in human genes, splicing reporter minigenes were derived from natural A5E and A3E exons which had substantial EST coverage of both alternative isoforms (Supplemental Data). Exon 9 of the human AGER (advanced glycosylation end product-specific receptor) gene has two alternative 5′ss separated by sequences containing overlapping FAS-hex3 ESS hexamers (Fig. 2C). Compared to the wildtype exon, a two-base mutation that disrupted these ESSs resulted in a substantial increase in usage of the intron-proximal 5′ss (Fig. 2C). Exon 3 of the human H2RSP (HAI-2 related small protein) gene has two alternative 3′ss, again separated by multiple FAS-hex3 ESS hexamers, and also undergoes exon skipping. Mutation of the ESS hexamers located between the alternative 3′ss had two effects, reducing the level of exon skipping and also increasing the relative usage of the intron-proximal 3′ss (Fig. 2D). Therefore, in this instance, ESSs function in regulation of both exon skipping and alternative 3′ss choice. These and similar results obtained for the IL17RE gene (Fig. S3) support the idea that ESSs located between alternative splice sites commonly regulate splice site usage in human genes.
The degree of conservation of ESE hexamers in the same sets of orthologous human-mouse A5E and A3E exons was also explored. The non-alignment-based COR method showed significant ESE conservation in the extension regions for both categories of exons (Fig. S2D). In interpreting these results on ESE conservation it should be kept in mind that for both classes of alternative splice site exons, some of the ESE conservation observed in the extension regions may reflect selection for exon inclusion rather than for regulation of alternative splice site usage. Furthermore, previous studies have indicated that ESE activity may often depend on position relative to the regulated splice site (e.g., Fairbrother et al., 2004; Graveley et al., 1998). Therefore, alignment-based metrics such as the number of conserved ESEs per 100 nt may be more appropriate for studying ESEs. Using this measure, we observed a higher density of conserved ESEs in the extension regions of A5Es than in the core regions, but no significant difference in ESE density between the core and extension regions of A3Es (Fig. S2C). These results mirror the experimental results described above which showed that ESEs have more pronounced effects on competing 5′ss than competing 3′ss (Fig. S1B, S1D), and suggest that ESEs may be commonly used to regulate splice site usage in endogenous alternative 5′ss exons.
Tethering of hnRNP A1 and SF2/ASF Mimics Effects of ESSs and ESEs
Both ESEs and ESSs are thought to commonly function through binding to specific trans-acting protein factors, whose level or activity may differ between cell types (Black, 2003). To test whether the effects on splice site selection observed above could be explained by recruitment of canonical ESE- and ESS-associated factors, fusion proteins with the phage MS2 coat protein were used. The MS2 RNA hairpin, which is bound with high affinity by the MS2 coat protein, was inserted into the competing splice site reporter constructs in the location used to test candidate splicing regulatory elements, and cells were co-transfected with expression constructs for the MS2 coat protein fused to either the ESE-binding SR protein SF2/ASF, the ESS-binding hnRNP A1 protein, or the glycine-rich domain of A1 (Fig. 3) (Del Gatto-Konczak et al., 1999). Co-transfection of MS2-SF2/ASF with the competing 5′ss-MS2 hairpin reporter resulted in increased usage of the intron-proximal 5′ss (Fig. 3A, lane 3) relative to mock-transfected controls. The MS2 fusions with either hnRNP A1 or its glycine-rich domain had the opposite effect, inhibiting use of the intron-proximal 5′ss (Fig. 3A, lanes 5 and 7). That these shifts in splice site usage result from specific binding of the fusion proteins to the MS2 hairpin is supported by the observation that much smaller shifts were observed with the control MS2Δ reporter that contains a single nucleotide deletion in the MS2 hairpin which essentially abolishes binding to coat protein (lanes 4, 6, 8 of Fig. 3A). This mutation had no effect on splice site usage in the absence of fusion protein (lanes 1, 2 of Fig. 3A). Performing similar experiments using the competing 3′ss reporter, fusions with hnRNP A1 or its glycine-rich domain again strongly inhibited the intron-proximal splice site, but the SF2/ASF fusion protein had little or no effect on 3′ss usage in this system (Fig. 3B).
Figure 3. Effects of tethering splicing factors between competing splice sites.
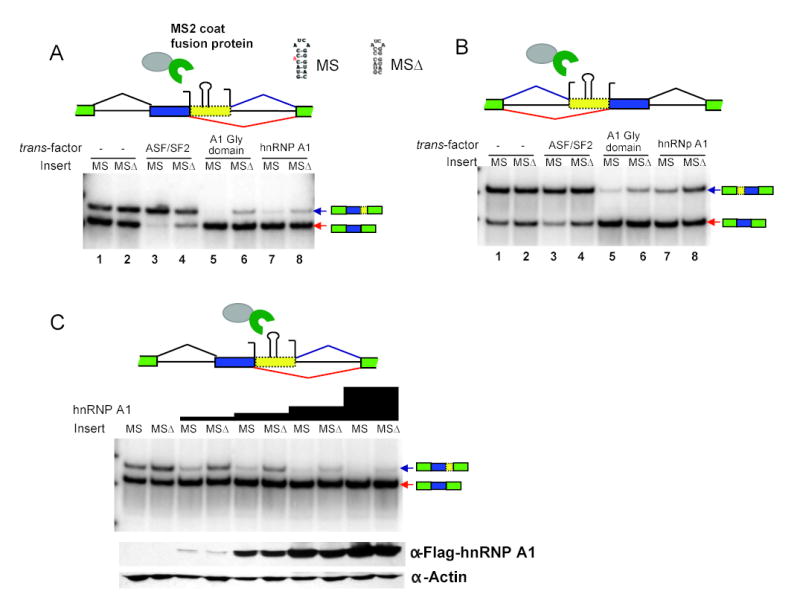
(A) Results of cotransfection of MS2-splicing factor fusion protein expression constructs with competing 5′ss reporter (as in Fig. 1B, 1C), using MS2 hairpin inserted in place of splicing regulatory element (- indicates mock-transfection). Sequences are shown for MS, an RNA hairpin bound with high affinity by MS2 coat protein; and MSΔ, a hairpin with 1 nt deletion which abolishes coat protein binding. Fusion proteins shown as gray oval (splicing factor) fused to green crescent (MS2 coat protein). Products of body-labeled RT-PCR of total RNA extracted from co-transfected cells using primers in flanking exons. (B) Competing 3′ss reporter with MS2 hairpin insert. Co-transfection and RT-PCR analysis as in panel A. (C) Results of transfecting increasing quantities of hnRNP A1-MS2 coat fusion protein expression construct (0.01 ug to 0.125 ug, proportional to height of grey bars above each lane) co-transfected with 0.5 ug of competing 5′ss reporter containing MS or MSΔ hairpin. RT-PCR as in panel A. The amount of fusion protein was detected by western blot using anti-Flag antibody (second panel from bottom). Bottom panel, loading controls of western blot using anti-actin antibody.
The strong effects of the hnRNP A1 fusions on both 5′ss and 3′ss usage in the presence of the MS2 hairpin mirrored the effects of similarly positioned ESSs (Fig. 1). The effects of SF2/ASF fusion protein on 5′ss but not 3′ss usage also paralleled the differential effects on 5′ss and 3′ss usage observed for many ESEs (Fig. S1). Thus, the effects on splice site selection mediated by many ESS and ESE elements could potentially be explained by direct recruitment of splicing factors of the hnRNP and SR protein families, respectively, and the effects of A1 on splice site choice are likely mediated through its glycine-rich domain.
Sequence Specificity of hnRNP and SR Protein Effects
The sequence-specific effects of the MS2 hairpin relative to the MS2Δ sequence were observed using ratios of fusion protein expression plasmid to splicing reporter plasmid of 0.05:1 and 0.1:1 for the competing 5′ss and 3′ss reporters, respectively. These ratios represent the lower ends of the ranges that gave robust splice site inhibition, and are substantially lower than those typically used in splicing factor/splicing reporter co-transfection experiments (Bai et al., 1999). When the ratio of MS2-A1 expression plasmid to 5′ss reporter plasmid was increased by up to 12.5-fold, a steady increase in non-specific inhibition of the intron-proximal splice site was observed, and at high levels of expression plasmid similar results were observed for the MS2 and MS2Δ reporters (Fig. 3C). Since the MS2Δ mutation essentially abolishes binding of MS2 coat protein (affinity reduced by > 3,000-fold) (Schneider et al., 1992), these data suggest that hnRNP A1 protein may have non-sequence-specific as well as sequence-specific effects on 5′ss usage. Additional support for this somewhat surprising conclusion comes from observing the effects of different concentrations of the A1 glycine-rich domain-MS2 coat fusion protein on splice site choice (Figure S4C). These data show that sufficiently high levels of this fusion protein, which completely lacks the canonical RNA-binding portion of the A1 protein, can inhibit the intron-proximal 5′ss in the presence of the mutant MS2 binding site. A similar loss in sequence specificity was observed when the ratio of MS2-SF2/ASF expression plasmid to competing 5′ss reporter was increased over a range of 12.5-fold (Fig. S4), suggesting that this factor may also have non-sequence-specific as well as sequence-specific effects on 5′ss usage.
It is well established that SF2/ASF and other SR proteins can promote the use of intron-proximal splice sites in several systems – either when added to in vitro splicing reactions (Fu et al., 1992; Ge and Manley, 1990; Krainer et al., 1990) or when over-expressed in vivo (Bai et al., 1999; Caceres et al., 1994). Conversely, addition or over-expression of hnRNP A1 has been observed to antagonize the activity of SR proteins in several systems, often inhibiting splicing at intron-proximal sites (Bai et al., 1999; Caceres et al., 1994; Fu et al., 1992; Mayeda and Krainer, 1992). These observations led to a model in which the levels of SR proteins relative to hnRNP A1 could modulate the selection of alternative splice sites (Mayeda and Krainer, 1992). However, these studies generally did not address the issue of sequence-specific binding of the target transcript by the splicing factors whose concentrations were manipulated.
Our results indicate that over-expression of SR or hnRNP splicing factors using typical expression protocols may often give rise to non-sequence-specific effects on splice site choice (Fig. 3C,Fig. S4), but that when splicing factors are expressed at more modest levels (as in Fig. 3A, B), or are expressed at endogenous levels (as in Figures 1 and 2), splice site selection is highly dependent on the sequences present between the competing splice sites. Thus, the non-specific effects observed with high levels of SR and hnRNP fusion protein expression constructs may simply be an artifact of over-expression. Taken together, the experimental and bioinformatic data described above support a model for splice site selection in which ESS sequences commonly mediate inhibition of intron-proximal decoy or alternative 5′ss or 3′ss, and ESEs often mediate an opposing effect on 5′ss selection. The set of ESSs and ESEs located between the splice sites could be used to tune the relative usage of a pair of alternative splice sites. Regulated splice site choice could then result from changes in the activities (expression levels, subcellular localization, modification state, etc.) of associated splicing regulatory proteins such as hnRNP and SR family proteins, each of which could potentially regulate a specific subset of A5E and A3E events depending on the precise ESS or ESE sequences present between the alternative splice sites. By contrast, if these factors simply promoted or inhibited use of all intron-proximal splice sites independent of sequence, such an activity would enable only a rather crude regulatory response to developmental or environmental cues. Thus, we propose that the region between alternative splice sites is critical for determining the level of usage of the alternative sites under different conditions or in different cell types.
Inhibition of Intron Retention by ESSs
Since ESSs can promote both exon skipping and alternative splice site usage, we next sought to determine whether ESSs can also regulate the remaining major class of alternative splicing events, intron retention. Though inserted into an intronic context in these experiments, we continue to refer to these elements as ESSs, keeping in mind that this designation implies nothing about their activity when located in an intronic context. For this purpose, we used a ‘multifunctional’ splicing reporter containing a pair of adjacent exons which are capable of splicing by multiple different pathways, leading to either: (i) inclusion of both exons with retention of the intervening intron (‘retained intron’ isoform); (ii) inclusion of both exons with splicing of the intervening intron (which we call the ‘fully spliced’ isoform because all pairs of splice sites are used); (iii) inclusion of the upstream exon with skipping of the downstream exon (‘single-skipped’ isoform); or (iv) exclusion of both exons (‘dual-skipped’ isoform). This combination of splicing events was found to occur in transcripts spanning exons 3 and 4 of the human NKIRAS2 (NFKB inhibitor interacting Ras-like 2) gene, from which a splicing reporter was derived (Fig. 4A). The multiple pathways of splicing which this pair of exons can undergo afforded an opportunity to assess the relative effects of ESSs on exon skipping and intron retention.
Figure 4. Role of ESS in regulation of intron retention.
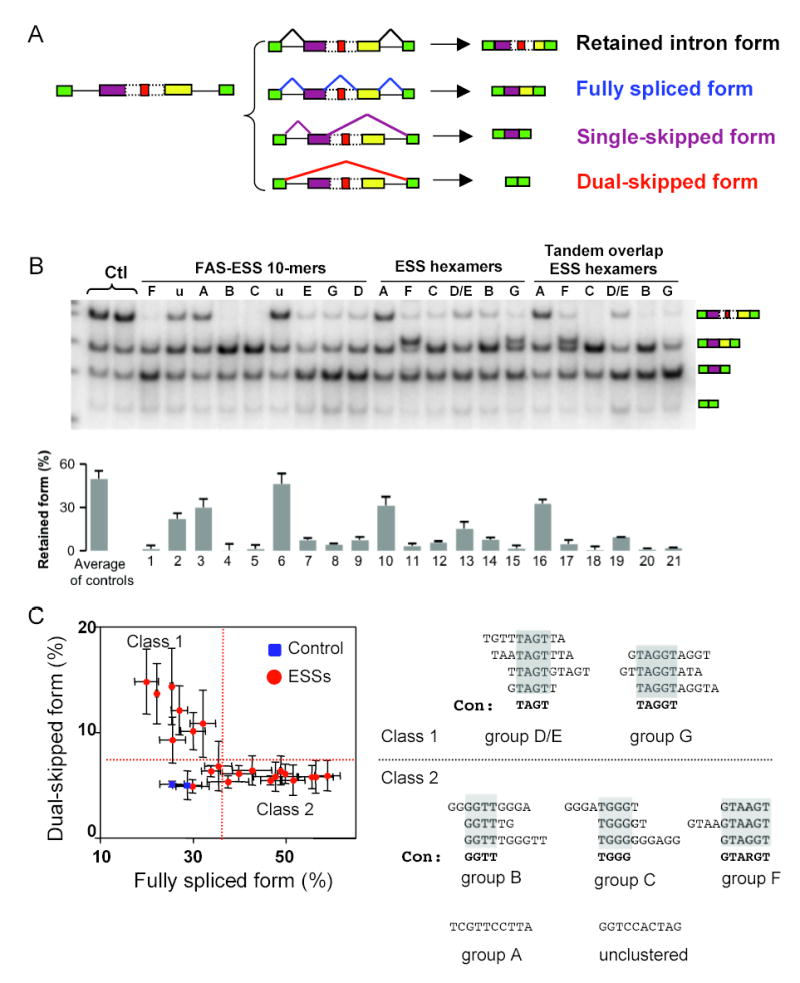
(A) Multifunctional intron reporter minigene and exon structures of four expected splicing products, ordered by decreasing size. Green boxes represent constitutive GFP exons; purple and yellow boxes represent variable exons; dashed box is retained intron; red box is inserted sequence. (B) Body-labeled RT-PCR of total RNA from cells transfected with intron retention minigene reporters inserted with control and ESS sequences (controls 1 and 2 listed in Table S5; other sequences in Table S3). Transfections were carried out as in Fig. 1C, in duplicate. Bottom panel shows the fraction of spliced product which represents the retained intron isoform in each sample (see Methods for details). Mean and range of control sequences is shown at left; error bars in remaining lanes indicate range of duplicated experiments. (C) Scatter plot showing fraction of fully spliced form and dual-skipped isoforms for ESSs (red dots) and control sequences (blue squares) in the experiment shown in panel B, showing range of dulicate values in each dimension. Red dotted lines illustrate division of ESSs into two activity classes: Class 1 ESSs increased level of dual-skipped isoform above level of horizontal red line; Class 2 ESSs increased level of fully spliced form above level of vertical red line; three ESSs clustered with control sequences (below both red lines). Sequences of tested ESS hexamers and decamers in each class are shown at right, grouped by sequence similarity, with similar segments indicated by shading. Consensus sequences and group names from (Wang et al., 2004) are listed below each cluster.
The same diverse panel of ESS sequences and controls was inserted into the retained intron of the NKIRAS2 minigene, and splicing was assayed by transient transfection followed by quantitative RT-PCR to measure relative changes in spliced isoform levels (Fig. 4B). The retained intron form was the most abundant isoform produced from both the native construct and following insertion with control sequences (Fig. 4B,Fig. S5). Notably, insertion of most ESSs substantially reduced the relative level of the retained intron form (Fig. 4B, top band; the three exceptions were the same three sequences which failed to significantly affect 3′ss selection in Fig. 1D), suggesting that sequences capable of inhibiting exon inclusion from an exonic location may generally have the ability to inhibit intron retention from an intronic location. Consistent with this idea, it has recently been reported that intronic binding sites for certain hnRNP factors can stimulate the in vitro splicing of pre-mRNAs containing artificially enlarged introns (Martinez-Contreras et al., 2006).
Although reduction in the level of the retained intron isoform was observed nearly universally across the set of ESSs studied, different ESSs affected the other isoforms in different ways. While almost all of the ESSs studied increased the levels of the single-skipped form (Fig. 4B andFig. S5), a scatter plot of the levels of the fully spliced and dual-skipped isoforms for the tested ESSs and controls indicated a curious mutually exclusive pattern (Fig. 4C), suggesting the presence of two distinct classes of ESSs with distinct effects on splicing pathways. One group of ESSs, which we call Class 1, increased the dual-skipped form relative to the controls, with little or no effect on levels of the fully spliced isoform, while ESSs in the other group, which we call Class 2, increased the levels of the fully spliced form, with little or no change in the dual-skipped isoform (Fig. 4C). Strikingly, no tested ESS significantly increased the levels of both of these isoforms or significantly reduced the level of either of these isoforms relative to the controls. The distinct effects of the two classes of ESSs in this reporter can potentially be explained by differing effects of the corresponding trans-factors on splice site recognition and exon definition interactions (see Supplemental Data andFig. S6).
Consistent with their distinct effects on splicing of the NKIRAS2 minigene, the sequences of Class 1 and Class 2 ESSs were generally quite distinct from each other (Fig. 4C, right hand side). Class 1 sequences all contained either the tetramer TAGT or the pentanucleotide TAGGT, which resembles the binding motif for hnRNP A1 (Burd and Dreyfuss, 1994). Class 2 sequences included most other ESSs, including a pyrimidine-rich group A sequence; an unclustered sequence; several G-rich and G/T-rich sequences containing GGTT and/or TGGG, many of which resemble the binding motif for hnRNPs F/H (Chen et al., 1999); and sequences containing GTARGT (R = A or G), which is similar to the 5′ss consensus/GTRAGT. As described previously, this latter group of sequences, in addition to having ESS activity, can also be recognized as 5′ss (Wang et al., 2004). The extra band that appears in Fig. 4B above that for the fully-spliced isoform derives from use of these sequences in place of the normal 5′ss of the upstream exon.
The pronounced effects of ESSs on intron retention in the NKIRAS2 reporter suggested that ESSs might commonly be involved in cases of endogenous regulated intron retention. It is exceedingly rare to observe skipping of both of the exons flanking a retained intron in endogenous human genes: in a database of over 1200 retained introns, we observed only one clear-cut case of dual-exon skipping based on available transcript data, which was the NKIRAS2 gene, used in the reporter above. Thus, in a typical intron retention situation where isoforms that retain or splice out the intron are desired, but dual exon skipping is not desired, regulation of intron retention by Class 2 ESSs might be preferred, since Class 1 ESSs would tend to induce undesired dual exon skipping. To explore this idea, we analyzed the frequencies of Class 1 and Class 2 ESS hexamers in a large dataset of alternatively retained introns, in comparison to constitutively spliced introns. Consistent with the above model for regulation of intron retention, we observed that retained introns have significantly lower frequencies of Class 1 ESSs and significantly higher frequencies of Class 2 ESSs than controls, at both the 5′ and 3′ ends of the intron (Fig. 5). Thus, although additional studies will be required to establish the roles of ESSs in specific retained introns, the data presented in Figures 4 and 5 suggest that the Class 2 subset of ESSs may play a common role in inhibiting intron retention in human genes.
Figure 5. Distribution of ESS hexamers in retained introns.
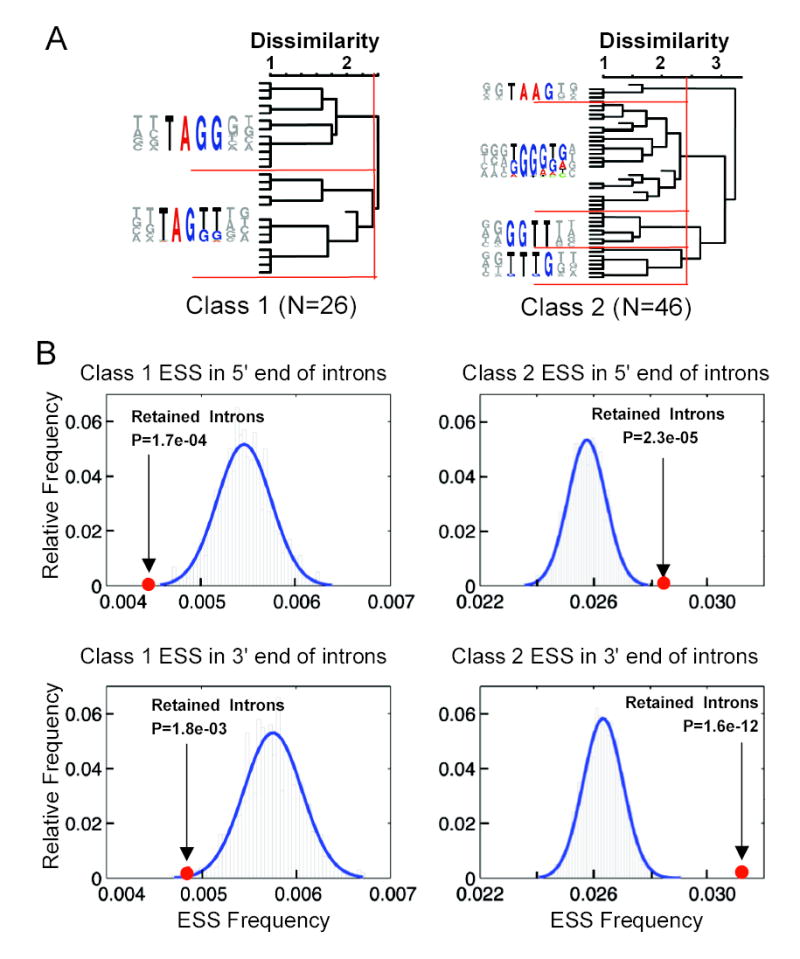
(A) Clusters of ESS hexamers with similar consensus sequences as Class 1 and Class 2 ESSs tested in Fig. 4. FAS-hex3 hexamers were classified based on these clusters; hexamers whose classification was unclear were omitted. (B) Distributions of Class 1 and Class 2 ESS hexamers in 5′ and 3′ ends of retained introns (5′ end includes 5′ half of intron, up to 200 bases; 3′ end includes 3′ half of intron, up to 200 bases). Histograms show ESS density in 2,000 sets of control constitutive introns with similar length and expected counts of ESS hexamers as retained introns. P-values were calculated based on a normal approximation to the distribution of the controls (blue curves).
Perspectives on the Context and Scope of ESS Activity in Control of Splicing
In many natural contexts, ESSs function cooperatively with each other or with other splicing regulatory elements, sometimes exhibiting highly complex interactions (Black, 2003; Matlin et al., 2005). For example, silencing of the brain-region-specific CI cassette exon (exon 19) of the glutamate NMDA R1 receptor (GRIN1) transcript is mediated by a pair of exonic TAGG elements which function together with a GGGG motif that overlaps the 5′ss (Han et al., 2005). Here, we have shown that a large and diverse set of ESS elements have consistent effects on splicing in a variety of different sequence contexts, including reporters based on human SIRT1 exon 6 and its flanking introns (Figs. 1, 3), human AGER exon 9 and H2RSP exon 3 and their flanking introns (Fig. 2C, 2D), and exons 3 and 4 and associated introns of human NKIRAS2 (Fig. 4). Thus, no specific flanking sequence context appears to be generally required for activity of most of these ESSs, although of course the magnitude of their effect on splicing is presumably modifiable by surrounding sequences and dependent on the expression of specific trans-acting splicing factors.
The set of ESSs studied here was obtained using an in vivo screen for sequences with the ability to inhibit inclusion of a constitutive exon. Our initial bioinformatic analyses of constitutive and alternative/skipped exons suggested that these ESSs are avoided in constitutive exons but may commonly be involved in control of exon skipping (Wang et al., 2004), and regulation of exon skipping by ESSs with sequences related to several of the FAS-ESS groups has been described previously (Chen et al., 1999; Del Gatto-Konczak et al., 1999). The bioinformatic and experimental analyses described here provide evidence that these ESSs are commonly involved in splice site definition of constitutive exons, and in the regulation of alternative 5′ss usage, and in the regulation of alternative 3′ss usage, and in the regulation of intron retention. These diverse functions of ESSs are summarized in Fig. 6. Thus, it appears that ESSs play important roles in control of all of the common types of alternative splicing that affect human genes.
Figure 6. General roles of ESSs in alternative splicing.
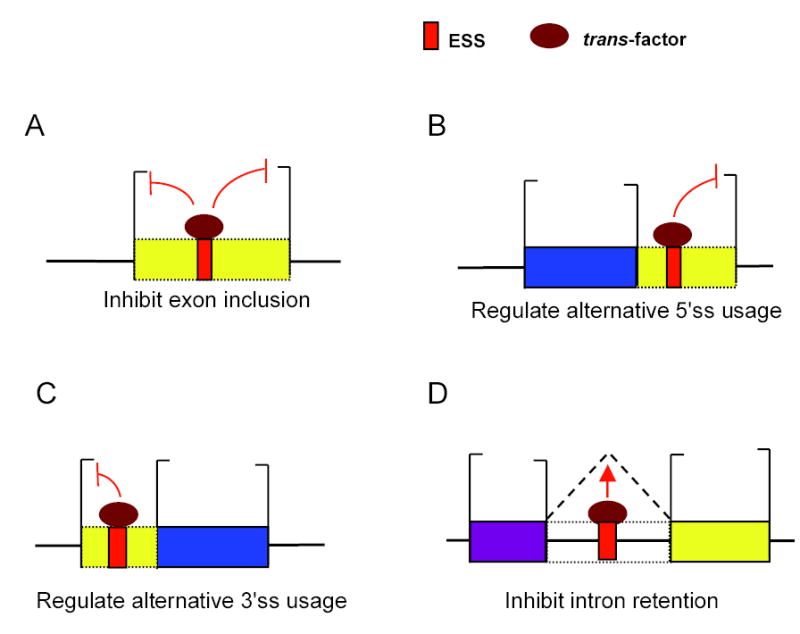
Model of general effects of ESSs on four common types of alternative splicing. Trans-factor represents a factor which binds to the ESS and mediates – directly or indirectly – the indicated effect on splicing. Summary of ESS effects: (A) inhibition of exon inclusion (based on previous bioinformatic and experimental studies – see text); (B) inhibition of intron-proximal 5′ss usage (supported by Figs. 1–3); (C) inhibition of intron-proximal 3′ss usage (supported by Figs. 1–3); (D) inhibition of intron-retention (supported by Figs. 4–5).
Experimental procedures
Reporter constructs
All the reporter systems were constructed with a backbone vector, pZW1, which contains a multicloning site between the two GFP exons (Wang et al., 2004).
To make reporters with competing 5′ss or competing 3′ss, the test exon (exon 6 of human SIRT1 gene, Ensembl ID: ENSG00000096717) together with portions of its flanking introns was inserted between two GFP exons, and the additional splice sites were constructed adjacent to the natural site. The ESS sequences were inserted between two competing splice sites by restriction enzyme digestion and ligation. The intron retention reporter was constructed using similar strategy.
To make reporters for the endogenous A5E and A3E events, the exons together with portions of their flanking introns were amplified by PCR, and inserted into pZW1. The mutant A5E and A3E reporters were generated by PCR with primers bearing point mutations designed to disrupt the ESSs as shown in Fig. 2. Additional details are provided in Supplemental Data.
Cell culture, transfection, RNA purification and RT-PCR
293 cells were cultured with D-MEM medium supplemented with 10% fetal bovine serum. Transfections were carried out with lipofectamine 2000 (Invitrogen) in 12-well culture plates.
Total RNA was purified from transfected cells using PURESCRIPT RNA isolation kit (Gentra Systems), follwed by DNase I treatment. The reverse transcription reaction was carried out using 2 μg total RNA with SupperScript III (Invitrogen). One tenth of the product from the RT reaction was used for PCR (20 cycles of amplification, with trace amount of α-32P-dCTP in addition to non-radioactive dNTPs). Quantification of splicing isoforms is described in Supplemental Methods.
Expression of MS2 coat fusion protein and western blotting
The expression constructs for fusion protein of MS2 coat protein (in pCI-MS2-NLS-FLAG vector) were gifts of Dr. R. Breathnach from Institut de Biologie-CHR (Del Gatto-Konczak et al., 1999). Varying amount of expression vectors were co-transfected with 0.5 μg of reporter construct in a 12-well culture plate, the RNA were purified and analyzed 24 hours after transfection as described above.
To detect hnRNP A1 expression by western blotting, the cells were harvested 24 hours after transfection. Total cell pellets were boiled in SDS-PAGE loading buffer for 5 min, and then separated by precast SDS-PAGE gel (4-20% gradient, from Bio-Rad). The fusion protein was detected with anti-FLAG antibody (monoclonal anti-FLAG M2 antibody from Sigma-Aldrich) and visualized with ECL kit (Amersham).
Exon and Intron Data Sets
Alignments of human and mouse cDNA and EST sequences to the human and mouse genomes were obtained from the UCSC Genome Browser (Karolchik et al., 2003). Constitutive exons (CEs), pairs of alternative 3′ss exons (A3Es), pairs of alternative 5′ss exons (A5Es), skipped exons (SEs) and retained introns (RIs) were categorized as in (Yeo et al., 2004). Additional filters were applied to minimize potential EST alignment artifacts (see Supplemental Data). Potential splice sites were scored using a maximum entropy model (Yeo and Burge, 2004) to generate datasets of CEs with and without upstream decoy 3′ss (or downstream decoy 5′ss). Decoy-dependent peaks of ESS density were most prominent in shorter introns (< 1kb), becoming weaker as intron length increased.
The above human and mouse exons were identified independently by using transcript data specific to each organism. Human/mouse orthologous A3Es, A5Es, SEs and CEs were identified based on the human-centric multiz (multiz8way) alignment obtained from the UCSC Genome Browser (Karolchik et al., 2003). To obtain datasets of high quality for analysis of conservation of splicing regulatory elements in A3Es and A5Es, we devised a procedure to obtain putative human/dog/mouse/rat orthologous A3Es and A5Es based on human sequences and multi-genome alignments (see Supplemental Data). A total of 1074 A5Es and 1318 A3Es passed the filtering procedure and are categorized as potential orthologous A5Es and A3Es in the human, dog, mouse and rat genomes.
Conservation of ESS and ESE Hexamers in A3E and A5E exons
To analyze the conservation of ESS and ESE hexamers in the A3E and A5E extension regions, we defined a measure called Conserved Occurrence Rate (COR):
where CORH and CORM are measures of conservation of human and mouse oligonucleotide sequence sets, respectively. CORH and CORM are defined as follows:
where the upper sum is taken over all i, j pairs such that ; and
where the upper sum is taken over all i, j pairs such that . Here, represents the number of occurrences of the jth FAS-hex3 ESS hexamer in the ith human exon region, and represents the number of occurrences of this hexamer in the corresponding region of the mouse ortholog of the ith human exon. In Fig. 2B, the regions under consideration were the regions between alternative 5′ss or 3′ss pairs in sets of orthologous A5E and A3E human/mouse exon pairs. The difference in occurrence counts are summed as indicated over all ESS hexamers and all pairs of orthologous exons. The COR for ESEs was defined similarly using the set of RESCUE-ESE hexamers. Note that this definition is ‘alignment-independent’ in that, in order to achieve the maximum COR value of 1.0 it is sufficient that the counts of the set of hexamers be the same in the corresponding human/mouse exon regions, but it is not required that these hexamers be aligned. This definition is closely related to alignment-based metrics such as the ‘Conservation Rate’ (Xie et al., 2005), but may be more appropriate for splicing regulatory elements which are relatively short and can function at various positions within an exon. For the background distribution, COR values were calculated for random control sets of hexamers that had exactly the same total number of occurrences as the ESSs (or ESEs) in the A5E and A3E extension region (see Supplemental Data).
Supplementary Material
Acknowledgments
We thank B. Blencowe and P. A. Sharp for helpful comments on the manuscript, R. Sandberg and M. Stadler for providing exon datasets, and R. Breathnach for providing fusion protein constructs.
References
- Bai Y, Lee D, Yu T, Chasin LA. Control of 3′ splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res. 1999;27:1126–1134. doi: 10.1093/nar/27.4.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. Embo J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Cascino I, Papoff G, De Maria R, Testi R, Ruberti G. Fas/Apo-1 (CD95) receptor lacking the intracytoplasmic signaling domain protects tumor cells from Fas-mediated apoptosis. J Immunol. 1996;156:13–17. [PubMed] [Google Scholar]
- Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Forrest ST, Barringhaus KG, Perlegas D, Hammarskjold ML, McNamara CA. Intron retention generates a novel Id3 isoform that inhibits vascular lesion formation. J Biol Chem. 2004;279:32897–32903. doi: 10.1074/jbc.M404882200. [DOI] [PubMed] [Google Scholar]
- Fu XD, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci U S A. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Mine M, Marsac C, Brivet M, Tazi J, Soret J. The SR protein SC35 is responsible for aberrant splicing of the E1alpha pyruvate dehydrogenase mRNA in a case of mental retardation with lactic acidosis. Mol Cell Biol. 2005;25:3286–3294. doi: 10.1128/MCB.25.8.3286-3294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. Embo J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt B, Koziell A, Poulat F, Wieacker P, Scambler P, Berta P, Gessler M. Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1 +/−KTS splice isoforms. Hum Mol Genet. 1998;7:709–714. doi: 10.1093/hmg/7.4.709. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Cavaloc Y, Stevenin J. Alternative splicing of intron 3 of the serine/arginine-rich protein 9G8 gene. Identification of flanking exonic splicing enhancers and involvement of 9G8 as a trans-acting factor. J Biol Chem. 2001;276:7850–7858. doi: 10.1074/jbc.M009510200. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Schneider D, Tuerk C, Gold L. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J Mol Biol. 1992;228:862–869. doi: 10.1016/0022-2836(92)90870-p. [DOI] [PubMed] [Google Scholar]
- Sugnet CW, Kent WJ, Ares M, Jr, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac Symp Biocomput. 2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


