Abstract
DNA methylation at CpG residues is closely associated with a number of biological processes during vertebrate development. Unlike the vertebrates, however, several invertebrate species, including the Drosophila, do not have apparent DNA methylation in their genomes. Nor have there been reports on a DNA (5-cytosine) methyltransferase (CpG MTase) found in these invertebrates. We now present evidence for two CpG MTase-like proteins expressed in Drosophila cells. One of these, DmMTR1, is a protein containing peptide epitopes immunologically related to the conserved motifs I and IV in the catalytic domain of the mammalian dnmt1. DmMTR1 has an apparent molecular mass of 220 kDa and, similar to mammalian dnmt1, it also interacts in vivo with the proliferating cell nuclear antigen. During interphase of the syncytial Drosophila embryos, the DmMTR1 molecules are located outside the nuclei, as is dnmt1 in the mouse blastocyst. However, DmMTR1 appears to be rapidly transported into, and then out of the nuclei again, as the embryos undergo mitotic waves. Immunofluorescent data indicate that DmMTR1 molecules “paint” the whole set of condensed Drosophila chromosomes throughout the mitotic phase, suggesting they may play an essential function in the cell-cycle regulated condensation of the Drosophila chromosomes. Through search in the genomic database, we also have identified a Drosophila polypeptide, DmMT2, that exhibits high sequence homology to the mammalian dnmt2 and the yeast CpG MTase homolog pmt1. The expression of DmMT2 appears to be developmentally regulated. We discuss the evolutionary and functional implications of the discovery of these two Drosophila proteins related to mammalian CpG MTases.
Keywords: epitope detection, early embryo, cell cycle, chromatin structure, database
The cytosine DNA methyltransferases, or C-methylating enzymes, are a family of conserved enzymes present in a variety of organisms (1). Of these, the vertebrate DNA (5-cytosine) methyltransferases (CpG MTases) specifically methylate CpG dinucleotides. Thus far, only one major form of CpG MTase abundantly expressed in the vertebrate somatic tissues has been identified, cloned, and extensively characterized (2–7). This protein, dnmt1, is essential for early embryonic development, as demonstrated by knockout mice experiments (8). The mammalian dnmt1 enzyme is approximately 1,620 aa long and consists of two distinct domains (Fig. 1). The C-terminal, catalytic domain of dnmt1 is of similar length as the bacterial C-methylating enzyme, and it shares with the latter a common architecture consisting of 10 conserved amino acid motifs, I through X, each carrying out specific functions (for references, see ref. 9) (Fig. 1).
Figure 1.
Conservation of CpG MTase-like proteins in eukaryotes. The general structural organization of different eukaryotic CpG MTase-like proteins are shown with their methylation activities indicated. Those of the bacterial C-methylating enzymes are included for comparison. Of the 10 conserved sequence motifs in the catalytic domains, only I and IV, which are relevant to the present study, are indicated. The boundaries of DmMTR1 and DmMT2 are unknown at the present time, and are represented with the dashed lines. For positions of the conserved motifs in DmMT2, see Fig. 7.
The N-terminal regulatory domain of the dnmt1 protein, which is absent from the bacterial enzymes, contains the replication foci-targeting sequence (10), two nuclear localization signals (NLSs) (10, 11), and a region interacting with proliferating cellular nuclear antigen (PCNA) (12), a protein involved in DNA replication and DNA repair (13, 14). These structural characteristics reflect the proposed involvement of the vertebrate dnmt1 proteins in various nuclear functions including mutagenesis, DNA repair, genomic imprinting, mammalian X chromosome inactivation (reviewed in refs. 15 and 16), and chromatin organization (17, 18).
Several other mammalian CpG MTase homologs have been identified recently (Fig. 1). These include dnmt2 (19–21), which lacks the N-terminal domain of dnmt1; dnmt3a and dnmt3b (22), both of which contain shorter N-terminal domains with little homology to that of dnmt1. These new CpG MTases all are expressed at very low levels in adult tissues. However, dnmt3a and dnmt3b messages are abundant in mouse embryonic stem (ES) cells (22). Of these, dnmt3a and dnmt3b are genuine CpG MTases, whereas the function of dnmt2 in CpG methylation has not been demonstrated.
In contrast to the vertebrates, the genomes of Drosophila, Caenorhabditis elegans, and yeast are deficient in m5C residues, at least at the resolution level of HPLC analyses (23–25). Evidence for the existence of CpG MTase-like proteins in these organisms also is lacking (see Discussion). A CpG MTase homolog, pmt1, expressed in yeast Schizosaccharomyces pombe cells has been cloned. This protein is 330 aa long and is most homologous to mammalian dnmt2 (ref. 26; Fig. 1). However, pmt1 lacks the ability to methylate DNA, most likely because of the proline-to-serine substitution in the conserved motif IV (27).
In this study, we have collected evidence for the existence of at least one CpG MTase homolog expressed in Drosophila cells. By immunobiochemical and immunocytological methods, we also have identified a polypeptide possessing several characteristics mimicking the mammalian dnmt1 enzymes.
Materials and Methods
Materials.
General molecular biology and biochemistry techniques are according to Sambrook et al. (28). Commercialized antibodies used include monoclonal anti-α-tubulin antibody (Sigma), mouse anti-PCNA (Santa Cruz Biotechnology), horseradish peroxidase-conjugated anti-rabbit (Zymed), and anti-mouse (Sigma), Cy3-conjugated anti-rabbit and Cy5-conjugated anti-mouse antibodies (Jackson ImmunoResearch). The DNA dyes, sytox and Hoechst 33258 are from Molecular Probes.
Preparation of Anti-Region I (RI) and Anti-Region IV (RIV) Antibodies.
Peptide antibodies were raised against two of the conserved regions, RI and RIV, of the mammalian CpG MTases. The two antigen sequences, RI and RIV, were identical to the human dnmt1 amino acids 1142–1156 or mouse dnmt1 amino acids 1145–1159, and to human dnmt1 amino acids 1217–1231 or mouse dnmt1 amino acids 1220–1234, respectively. The commercially synthesized peptides were individually coupled covalently to BSA (Sigma) as described (29) and then used as antigens. The antibodies were purified by the respective peptide conjugation to the epoxy-activated Sepharose 6B column (Amersham Pharmacia) (30). The peak fractions detected by Bio-Rad Protein Assay kit were pooled and dialyzed against PBS buffer at 4°C. The preimmune serum was processed in a similar way as described above and used in control experiments.
Construction of Glutathione S-Transferase (GST) Hybrids and Purification of the Fusion Proteins.
A 1.5-kb DNA fragment containing the catalytic domain of human CpG MTase cDNA, from 3629 to 5130, was amplified by PCR. The fragment was cloned into the pGEX-4T-2 vector (Amersham Pharmacia) in-frame to the GST gene resulting in the plasmid pGST-3′hMT. A fragment containing simian virus 40 NLS sequence was inserted in front of the 3′hMT to generate plasmid pGST-NLS-3′hMT. The fusion polypeptide was induced by isopropyl β-dD-thiogalactoside, and purified as described by the manufacturer. GST protein fused to the NLS-3′hMT was removed by treatment with thrombin (Amersham Pharmacia).
Western Blotting and Immunoprecipitation.
The extract of Drosophila Oregon R embryos was homogenized in the lysis buffer (8 M urea/1 mM PMSF/10 μg/ml pepstatin/2 mM each of leupeptin and aprotinin/0.1 mM DTT). For extract of the Schneider cell line 2 (SL2), the cells were lysed with TNGEK buffer (50 mM Tris⋅HCI, pH 8.5/1% NP-40/10% glycerol/0.4 M KCl/25 mM EDTA/2 mM PMSF/1 μg/ml of pepstatin and aprotinin/2 μg/ml leupeptin). The supernatant was collected upon centrifugation.
For immunoprecipitation experiments, the embryo extract was prepared in RIPA buffer (50 mM Tris⋅HCI, pH 7.4/1% NP-40/0.25% SDS/150 mM NaCl/1 mM EGTA/1 mM PMSF/1 μg/ml each of pepstatin, leupeptin, and aprotinin), and all reactions were incubated at room temperature. For immunocomplex analysis, the NET buffer (same as the RIPA buffer except that the concentration of NP-40 is 0.1%) was used instead, and the incubations were proceeded at 4°C. Hybridizing bands of Western blotting of the reaction products were visualized by using the ECL Western blotting detection system (Amersham Pharmacia).
Immunocytochemistry.
Zero to two-hour Drosophila Oregon R embryos were collected. Taxol pretreatment, fixation, and devitellinization were done as described in ref. 31. The primary antibodies used were anti-RI or anti-RIV along with monoclonal anti-α-tubulin antibody, and the secondary antibodies were Cy3-conjugated anti-rabbit and Cy5-conjugated anti-mouse antibodies. In most cases, the DNA dye sytox was added to a final concentration of 100 nM for 10 min before the last wash. The embryos in Fig. 4A were stained with 1 μg/ml of Hoechst 33258. Finally, the embryos were examined in Zeiss fluorescent microscope or confocal microscope and processed by using the Adobe photoshop program.
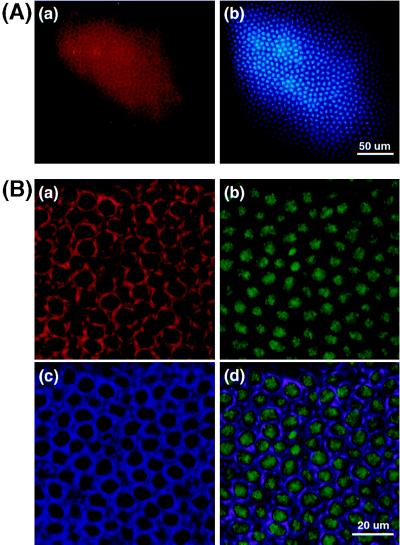
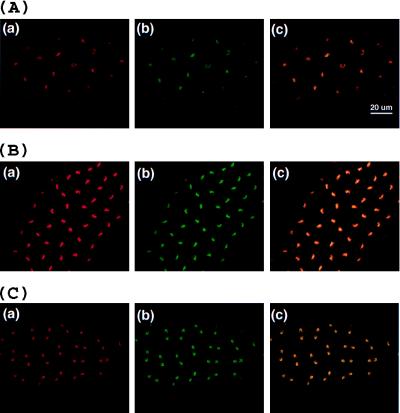
Figure 4.
Cellular distribution of DmMTR1 in Drosophila embryos at interphase. (A) Low magnification picture of a stage 14 embryo at interphase. (a) DmMTR1 (red) recognized by anti-RI; (b) DNA (blue) stained with Hoechst 33258. (B) High magnification of a stage 13 embryo at interphase. (a) DmMTR1 (red) recognized by anti-RIV; (b) DNA (green) stained with sytox; (c) α-tubulin (blue) recognized by monoclonal anti-α-tubulin antibody; (d) a merged image of DmMTR1, DNA, and α-tubulin.
Search for CpG MTase Homolog(s) from Drosophila Database.
The conserved motifs (I, II, IV, VI, VIII, IX, and X) of human dnmt1, mouse dnmt1, and S. pombe pmt1 proteins were run through the Drosophila sequence databank (BDGP, Berkeley Drosophila Genome Project, http://www.fruitfly.org) for blast (32) and pattern similarity search. The positive hits then were examined. Drosophila cosmids were obtained from the Medical Research Center Human Genome Mapping Resource Center at Hinxton, Cambridge, U.K. Alignment was predicted by clustal w program (33).
PCR.
Two primers (5′-GGAATTCACAGGCGCGA-3′ and 5′-CGGCTCGAGTCCATGACCAG-3′) were used to amplify a specific fragment DM from Drosophila Oregon R genomic DNA. This fragment encodes the region of DmMT2 from amino acids 124–222 (see Fig. 7). The DM fragment was cloned into pBluescript KS(−) vector (Strategene) and sequenced.
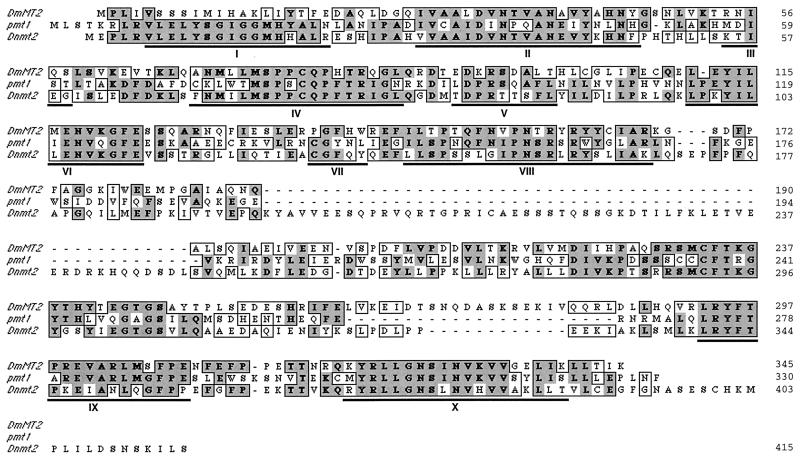
Figure 7.
Sequence alignment of DmMT2 with yeast pmt1 and mouse dnmt2. The ORF of DmMT2 is shown and aligned with yeast pmt1 (GenBank accession no. X82444) and mouse dnmt2 (GenBank accession no. AF012129). The identical amino acids are shown in dark gray boxes, and the homologous residues are marked in light gray boxes. The regions underlined indicate the conserved motifs from I to X.
Northern Blot Analysis.
Expression of DmMT2 was analyzed by Northern blot technique (28). Thirty micrograms of total RNA isolated from Drosophila embryos, larvae, and adult flies was loaded onto a 1% formaldehyde agarose gel. After electrophoresis, the gel was blotted and hybridized with the putative coding region of cloned DmMT2 labeled with 32P.
Results
A 220-kDa Drosophila Protein with Epitopes of the Conserved RI and RIV.
To see whether any protein with similar properties as the vertebrate CpG MTases is expressed in Drosophila cells, we first synthesized oligopeptides homologous to the conserved RI and RIV of the human and mouse CpG MTase dnmt1. Of the two, RI interacts with cofactor adenosyl methionine, whereas RIV has an invariant cysteine, which is part of the catalytic site, among the known CpG MTases (9, 26). Antibodies raised against these oligopeptides, anti-RI and anti-RIV, were purified and used as probes for blots containing extracts from Drosophila embryos and from the Drosophila SL2 cell line. As exemplified in Fig. 2A for anti-RI, both antibodies specifically recognize the C-terminal domain of human dnmt1 (NLS-3′hMT) expressed in bacteria as a GST fusion polypeptide, but not GST.
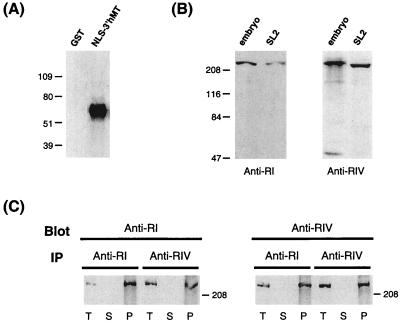
Figure 2.
Western blot analysis of Drosophila CpG MTase-like proteins. (A) Hybridization of GST or NLS-3′hMT polypeptide with anti-RI. The generation of the NLS-3′MT is described in Materials and Methods. The numbers on the left indicate the molecular masses (kDa) of markers. (B) Hybridization of extracts from Drosophila embryos or the Drosophila SL2 cell line. The probes used are anti-RI (Left) and anti-RIV (Right). The approximately 47-kDa band in the “embryo” lane of the right panel is not reproducible and could be the result of degradation. (C) Cross-immunoprecipitation (IP) and Western blot analysis using anti-RI and anti-RIV. Drosophila embryo extract was immunoprecipitated with anti-RI or anti-RIV, and then hybridized with anti-RIV or anti-RI, respectively. Samples on the blots include the total extract (T), supernatants after IP (S), and precipitates from IP (P).
Interestingly, each antibody also detects one major band on the Western blot of the Drosophila extracts (Fig. 2B). No band could be detected with the use of preimmune sera (data not shown). The molecular mass of the band, 220 kDa, is similar to that of the vertebrate dnmt1. Furthermore, the 220-kDa bands detected by anti-RI and anti-RIV most likely are derived from the same protein, as demonstrated by immunoprecipitation and cross-probing experiments of Fig. 2C. These data suggest that a 220-kDa protein is indeed expressed in Drosophila cells, and it contains two epitopes similar to the conserved RI and RIV, respectively. Because the cDNA has not been identified and cloned yet, we tentatively refer to this protein as DmMTR1, or Drosophila melanogaster CpG MTase-related protein 1.
DmMTR1 Interacts with PCNA in Vivo.
The mammalian dnmt1 associates with PCNA in vivo during S phase, and this complex association has been suggested to be involved in both DNA replication and DNA repair. To see whether DmMTR1 also physically interacts with PCNA in vivo in Drosophila cells, coimmunoprecipitation and cross Western blot hybridization experiments have been carried out. Drosophila embryo extract first was immunoprecipitated with the anti-RI or anti-RIV antibody. The total extract before precipitation (Fig. 3, lanes T), the supernatant (Fig. 3, lanes S), and precipitates (Fig. 3, lanes P) then were electrophoresed, blotted, and hybridized with anti-PCNA antibody.
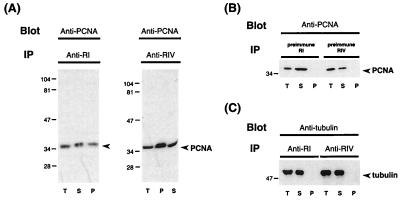
Figure 3.
Interaction of Drosophila DmMTR1 with PCNA. (A) Drosophila embryo extract was immunoprecipitated (IP) with anti-RI (Left) or anti-RIV (Right), and then hybridized on Western blot with anti-PCNA. (B) Control experiment with Drosophila embryo extract immunoprecipitated (IP) with preimmune sera, and then hybridized with anti-PCNA. (C) Blot hybridization of the same samples in A with antitubulin. Samples on the blots include the total extract (T), supernatants after IP (S), and precipitates from IP (P).
As seen in Fig. 3A, a band of similar molecular mass as the mammalian or Drosophila PCNA, 36 kDa, could be detected in either precipitate. This band was not precipitated by the preimmune sera (Fig. 3B). The specificity of the immunoprecipitation reaction was further demonstrated by blot hybridization of the same samples with antitubulin antibody. The 50-kDa tubulin band was seen only in the total extract and the supernatant, but not in the precipitate (Fig. 3C). We also have immunoprecipitated the Drosophila extract with anti-PCNA, and then blot-hybridized the samples with anti-RI or anti-RIV (data not shown). The result supports that deduced from the experiments of Fig. 3. That is, similar to mammalian dnmt1, the Drosophila DmMTR1 also associates physically with PCNA in vivo in Drosophila cells.
Chromatin Association of Drosophila DmMTR1.
Both antibodies, anti-RI and anti-RIV, were used to determine the distribution of DmMTR1 in Drosophila embryos. The staining patterns by the two antibodies are similar, if not identical, which supports the notion that they recognize the same protein.
As exemplified in Figs. 4–6, the cellular distribution of DmMTR1 in Drosophila early embryos is cell cycle-dependent. During interphase, DmMTR1 is present mainly in the cytoplasm, but not in nuclei, as observed for stage 13 and 14 embryos (Fig. 4). During mitosis, DmMTR1 becomes colocalized with DNA of the condensed mitotic chromosomes (Fig. 5). This DNA colocalization of DmMTR1 actually begins in prophase and persists until the end of mitosis (Fig. 6).
Figure 6.
Colocalization of DmMTR1 with chromosomal DNA during mitosis. DmMTR1 (red) is recognized by anti-RIV in a (A–C). DNA (green) is counterstained with sytox in b (A–C). The superimposed images of DmMTR1 and DNA are shown in c (A–C). (A) Prophase, a stage 11 embryo. (B) Metaphase, a stage 12 embryo. (C) Anaphase, a stage 11 embryo.
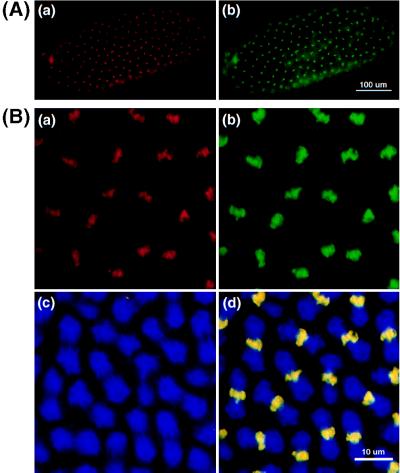
Figure 5.
Colocalization of DmMTR1 with DNA of metaphase chromosomes. (A) Low magnification of metaphase chromosomes of a stage 10 embryo. (B) High magnification of a stage 12 embryo. DmMTR1 (red) is stained with anti-RIV in Aa and Ba. DNA (green) is counterstained with sytox in Ab and Bb. The mitotic α-tubulin (blue) structure is shown in Bc. The superimposed image of DmMTR1, DNA, and α-tubulin is shown in Bd.
In the syncytial Drosophila embryos, the nuclear division is not completely synchronous. Divisions of cells in the two poles are initiated earlier than those at the equator. We have observed embryos undergoing the gradient of mitotic divisions (data not shown).
Identification of DmMT2, a Drosophila Homolog of Mammalian dnmt2 and Yeast pmt1.
With the use of conserved motifs (I, II, IV, VI, VIII, IX, and X) of human dnmt1, mouse dnmt1, and yeast S. pombe pmt1, we carried out a blast and pattern search and found two positive hits in Drosophila homologous to motif VIII of the yeast pmt1 protein. These hits are 19C7T and 87F11T, two almost identical sequence-tagged site sequences determined in the European Mapping Project. Of the two, 19C7T encodes an ORF of 114 aa. Appropriate DNA primers were designed according to sequences common to 19C7T and 87F11T and used to amplify a fragment, DM, from Drosophila genomic DNA. As described in the Drosophila database, 19C7 and 87F11 are located at loci 56D and 33C, respectively. Southern blot result using DM as the hybridization probe suggests that it is most likely single-copy in the genome (data not shown). To further confirm it, 23 cosmids previously mapped at 33C and 56D were analyzed by Southern blot. The DM fragment recognizes only 51C2, 87F11, and 102B1, all three of which are located at 33C, but not the other cosmids. This result suggests that the information regarding 19C7T in the database is incorrect.
We further sequenced DNA regions outside of DM in cosmid 102B1. The sequence including DM exhibits an ORF spanning about 1 kb encoding a polypeptide 345 aa long (Fig. 7). Surprisingly, this ORF contains sequences similar to the 10 conserved motifs of C-methylating enzymes. Furthermore, it is closer to mammalian dnmt2 and yeast pmt1 than to mammalian dnmt1. DmMT2 is approximately 50% homologous to both pmt1 and dnmt2. The homology covers not only the 10 motifs, but it extends to the whole dnmt2 and pmt1 sequences (Fig. 7). On the contrary, only 20% homology exists when any of these three proteins is compared with the catalytic domain of dnmt1 (data not shown). Therefore, we named this gene DmMT2.
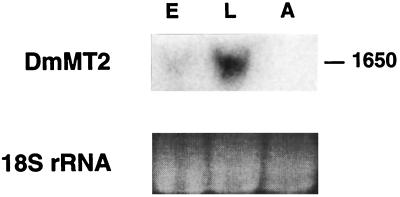
DmMT2 also is expressed. As shown in Fig. 8, Northern blot analysis indicates that DmMT2 is transcribed to generate a 1,650-nt RNA. The RNA species is most abundant in larvae, but only weakly expressed in embryos and undetectable at the adult stage. Thus, the expression of DmMT2 is developmentally regulated.
Figure 8.
Northern blot analysis of Drosophila DmMT2 RNA. DmMT2 expression in total RNAs of Drosophila embryos (lane E), larvae (lane L), and adult flies (lane A) is analyzed. The ethidium bromide staining of the Drosophila 18S rRNA is shown (Lower) as the standard.
Discussion
Although DNA methylation at CpG dinucleotides is a general phenomenon of eukaryotic genomes, methylated CpG residues have not been found in several well-known genetic model systems, such as the yeast, C. elegans, and Drosophila (23–25). Evidence for expression of CpG MTase(s) in these organisms also has been lacking. Our data show, however, that at least two proteins with characteristics similar to the mammalian CpG MTases are indeed expressed in Drosophila. These findings further suggest that the mammalian CpG MTases very likely carry out essential functions not requiring the enzymatic properties.
One of the CpG MTase-related Drosophila proteins we have identified is DmMTR1 (Fig. 1). Its relatively large molecular mass (220 kDa) is similar to that of the vertebrate dnmt1 enzymes (Fig. 2B). Cross-immunoprecipitation showed that DmMTR1 contains both the RI and RIV epitopes (Fig. 2C). Furthermore, just like the mammalian dnmt1, DmMTR1 also interacts physically with PCNA in vivo (Fig. 3). The immunobiochemical data were obtained independently with two different antibodies directed against the epitopes designed according to the conserved motifs, RI and RIV, respectively, of the mammalian dnmt1. These results together with the immunohistochemical data of Figs. 4–6 (see discussions later) suggest that DmMTR1 could be the Drosophila homolog of the mammalian dnmt1. However, a firm proof of this possibility awaits cloning of DmMTR1.
Immunofluorescent staining experiments of early Drosophila embryos revealed two unique cytological characteristics of DmMTR1. First, similar to a few eukaryotic proteins, including the yeast SWI5 (34), Drosophila pendulin (35), and Drosophila lodestar (36), DmMTR1 exhibits a cell cycle-specific switch of its locations in the cytoplasm and nucleus. It is cytoplasmic during the interphase (Fig. 4), but past interphase it exists only in the nuclei on the condensed chromosomes (Figs. 5 and 6). It is interesting to note here that the mouse CpG MTase dnmt1 also has specific patterns of cellular localization in cell culture and at different stages during early mouse embryo development (10, 37). In particular, in mouse blastocyst, which is probably equivalent developmentally to the Drosophila stage 13–14 embryo, the dnmt1 protein is also cytoplasmic in most of the presumably interphase cells (37). The cellular distribution patterns of DmMTR1 in Drosophila embryos of stages earlier than 12, and their cell cycle dependence awaits further detailed analysis.
Because the nuclear membranes and their associated nuclear pores remain essentially intact throughout different cell-cycle stages of the syncytially dividing nuclei, the compartment-specific localization of DmMTR1 could be the result of rapid resynthesis and degradation of the DmMTR1 molecules. Alternatively, rapid trafficking of the protein molecules across the nuclear membrane may be responsible for the observed switch of DmMTR1 localizations during the cell cycle progression. This model is supported, first, by the presence of the NLS as well as nuclear export sequences (NESs) in the mammalian CpG MTases. Two NLSs have been mapped within the mammalian dnmt1 (10, 11), and it is likely that DmMTR1 also contains such a signal. More interestingly, we have noticed the presence of NESs (38) in the mammalian dnmt1, dnmt2, and dnmt3 sequences (Fig. 9). The finding of NESs in the mammalian proteins suggest that cooperative functioning of NLSs and NESs may increase the efficiency for rapid nuclear trafficking of the mammalian CpG MTases as well as the related Drosophila protein DmMTR1. However, for either scenario described above, the signals directing these processes must be very active. Whether these signals are compartment-specific and/or regulated by cell cycle-specific events awaits future investigation.
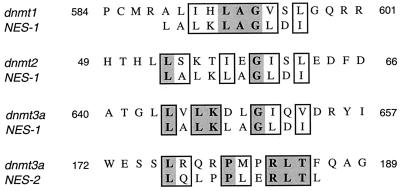
Figure 9.
Hypothetical NES in CpG MTase proteins. Two NESs, LALKLAGLDI and LQLPPLERLTL, have been identified (38). Among the mammalian MTase proteins dnmt1, dnmt2, and dnmt3a examined, either one of the NESs could be found in all of them.
Perhaps the most striking feature of DmMTR1 is its apparently uniform distribution on condensed chromatin during mitosis (Figs. 5 and 6). No such phenomenon has been reported for the mammalian dnmt1. There are quite a few nonhistone proteins known to be associated with condensed chromosomes (ref. 39 and references therein). However, of these, none is distributed throughout the chromosomes. In previous studies of Drosophila, only barren, a 70-kDa protein required for sister chromatid segregation in mitosis, is known to “paint” a major portion of the condensed chromosomes during mitosis (40). The chromosomal association of barren was proposed to be required for its function in mitotic sister chromatid segregation. DmMTR1 may be involved in a similar function, as well as in other activities during the mitotic phase.
It is also surprising that Drosophila encodes a protein, DmMT2, homologous to both mammalian dnmt2 and yeast pmt1. The alignment analysis of Fig. 7 suggests that the three proteins have originated from a common ancestor of the three species. The high sequence homology further suggests functional constrain(s) on the divergence rates of the three members.
So, what are the functions of DmMTR1 and DmMT2? First, one or both of them could be capable of methylating CpG dinucleotides, an activity that may have escaped from previous studies. Although it generally is accepted that Drosophila genome is void of methylated nucleotides, there was a report of very low level of m5C of Drosophila DNA as detected by a sensitive HPLC method (41). Such a methylation process may have very short duration time in Drosophila cells, for instance, only when the enzyme is associated with the chromatin DNA during mitosis. It should be noted here that both close members of DmMT2, the mammalian dnmt2 and yeast pmt1, lack methylation activities in vitro (21, 26).
Finally, DmMTR1 may play an essential function in chromosome condensation in Drosophila cells. Although other Drosophila chromosomal proteins, e.g., HP1, function to condense specific regions of the chromosomes such as the heterochromatin (42), DmMTR1 appears to act on the whole set of chromosomes during the mitotic phase of the embryos. In mammals, the methylated CpG residues preferentially bind certain proteins, including MeCP1 and MeCP2, an interaction proposed to be partially responsible for the observed repression of transcription (for references, see ref. 43). Indeed, it was recently demonstrated that MeCP2 can physically associate with histone deacetylases (HDACs) in vitro and in vivo (18, 45). HDACs capable of removing acetyl groups from the histones and causing chromatin condensation also are found in Drosophila (44). DmMTR1 may provide certain chromatin condensation function as the mammalian m5CpG/MeCP complexes. The exact roles of DmMTR1 and DmMT2 in chromosome condensation and other cellular activities will be better defined upon future molecular and genetic studies.
Acknowledgments
We thank Der-Hwa Huang and Ms. Y. L. Chang for their patience and technical advice on the embryo staining, Henry Sun for the Drosophila strains, Jerry Juang and National Health Research Institute for letting us use the Leica confocal microscope, and Roger Adams for providing MTase-specific antibodies at the beginning of the project. We are also grateful to Der-Hwa Huang, Henry Sun, Tao Hsieh, Cheng-Ting Chien, and our lab colleagues for constantly providing helpful discussions and suggestions. This research has been supported by the Academia Sinica and the National Science Council of Taipei, Taiwan.
Abbreviations
- CpG MTase
DNA (5-cytosine) methyltransferase
- PCNA
proliferating cellular nuclear antigen
- ES
embryonic stem
- RI
region I
- RIV
region IV
- GST
glutathione S-transferase
- NLS
nuclear localization signal
- NES
nuclear export sequence
Note Added in Proof
We have identified the cDNA and gene encoding a Drosophila protein related to a recently reported mammalian DNA demethylase or dMTase (unpublished work).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF185647).
References
- 1.Jost J P, Saluz H P, editors. DNA Methylation: Molecular Biology and Biological Significance. Basel: Birkhauser Verlag; 1993. [Google Scholar]
- 2.Bestor T H, Laudano A, Mattaliano R, Ingram V. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 3.Yoder J A, Yen R-W, Vertino P M, Bestor T H, Baylin S B. J Biol Chem. 1996;271:31092–31097. doi: 10.1074/jbc.271.49.31092. [DOI] [PubMed] [Google Scholar]
- 4.Yen R-W, Vertino P M, Nelkin B D, Yu J J, El-Deiry W, Cumaraswamy A, Lennon G G, Trask B J, Celano P, Baylin S B. Nucleic Acids Res. 1992;20:2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramchandani S, Bigey P, Szyf M. Biol Chem. 1998;379:535–540. doi: 10.1515/bchm.1998.379.4-5.535. [DOI] [PubMed] [Google Scholar]
- 6.Deng J, Szyf M. J Biol Chem. 1998;273:22869–22872. doi: 10.1074/jbc.273.36.22869. [DOI] [PubMed] [Google Scholar]
- 7.Tajima S, Tsuda H, Wakabayashi N, Asano A, Mizuno S, Nishimori K. J Biochem. 1995;117:1050–1057. doi: 10.1093/oxfordjournals.jbchem.a124805. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 9.Klimasauskas S, Kumar S, Roberts R J, Cheng X. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 10.Leonhardt H, Page A W, Weier H-U, Bestor T H. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Oakeley E J, Sun L, Jost J-P. Nucleic Acids Res. 1998;26:1038–1045. doi: 10.1093/nar/26.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang L S, Ian H I, Koh T W, Ng H H, Xu G, Li B F. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 13.Fairman M P. J Cell Sci. 1990;95:1–4. doi: 10.1242/jcs.95.3.1. [DOI] [PubMed] [Google Scholar]
- 14.Shivji M K K, Kenny M K, Wood R D. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 15.Yoder J A, Bestor T H. Biol Chem. 1996;377:605–610. [PubMed] [Google Scholar]
- 16.Laird P W, Jaenisch R. Annu Rev Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 17.Bestor T H. Nature (London) 1998;393:311–312. doi: 10.1038/30613. [DOI] [PubMed] [Google Scholar]
- 18.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 19.Yoder J A, Bestor T H. Hum Mol Genet. 1998;7:279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- 20.Okano M, Xie S, Li E. Nucleic Acids Res. 1998a;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyngaert I V den, Sprengel J, Kass S U, Luyten W H M L. FEBS Lett. 1998;426:283–289. doi: 10.1016/s0014-5793(98)00362-7. [DOI] [PubMed] [Google Scholar]
- 22.Okano M, Xie S, Li E. Nat Genet. 1998b;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 23.Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- 24.Proffitt J H, Davie J R, Swinton D, Hattman S. Mol Cell Biol. 1984;4:985–988. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson V J, Johnson T E, Hammen R F. Nucleic Acids Res. 1986;14:6711–6719. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson C R M, Bartlett R, Nurse P, Bird A P. Nucleic Acids Res. 1995;23:203–210. doi: 10.1093/nar/23.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinarbasi E, Elliott J, Hornby D P. J Mol Biol. 1996;257:804–813. doi: 10.1006/jmbi.1996.0203. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F M, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Pagano M, editor. Cell Cycle: Materials and Methods. Berlin: Springer; 1996. pp. 271–280. [Google Scholar]
- 30.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 31.Foe V E. Development (Cambridge, UK) 1989;107:1–22. [PubMed] [Google Scholar]
- 32.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgings D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasmyth K, Adolf G, Lydall D, Seddon A. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- 35.Küssel P, Frasch M. J Cell Biol. 1995;129:1491–1507. doi: 10.1083/jcb.129.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girdham C H, Glover D M. Genes Dev. 1991;5:1786–1799. doi: 10.1101/gad.5.10.1786. [DOI] [PubMed] [Google Scholar]
- 37.Mertineit C, Yoder J A, Iaketo T, Laird D W, Trasler J M, Bestor T H. Development (Cambridge, UK) 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- 38.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 39.Bhat K M, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. Development (Cambridge, UK) 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 40.Bhat M A, Philp A V, Glover D M, Bellen H J. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- 41.Patel C V, Gopinathan K P. Anal Biochem. 1987;164:164–169. doi: 10.1016/0003-2697(87)90381-2. [DOI] [PubMed] [Google Scholar]
- 42.Pak D T, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 43.Nan X, Javier Campoy F, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 44.Johnson C A, Barlow A L, Turner B M. Gene. 1998;221:127–134. doi: 10.1016/s0378-1119(98)00435-1. [DOI] [PubMed] [Google Scholar]
- 45.Jones P L, Jan Veemstra G C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]