The Informatics for Integrating Biology and the Bedside (i2b2) is one of the sponsored initiatives of the NIH Roadmap National Centers for Biomedical Computing (http://www.bisti.nih.gov/ncbc/). One of the goals of i2b2 is to provide clinical investigators broadly with the software tools necessary to collect and manage project-related clinical research data in the genomics age as a cohesive entity – a software suite to construct and manage the modern clinical research chart.
Maintaining relationships between terms in multiple vocabularies is a vital activity for any organization attempting to support the integration of data coming from multiple sources. In a clinical research setting, in fact, this activity becomes even more significant. Combined laboratory and clinical data from diverse systems, with semantically related codes, terms and concepts, can help transform genomic knowledge into the practice of healthcare. Unless these vocabulary relationships are defined, however, the potential for considerable insight is lost. Maintaining these relationships, then, becomes a priority.
Vocabulary mapping is a process of specifying and maintaining relationships between terms in multiple vocabularies. One vocabulary, designated as master becomes the classification scheme for the other subsidiary vocabularies.1 In order to use a system like this, there must be some way of maintaining freedom and control over the master vocabulary, while still maintaining the integrity of the mappings to each source. Though, there is no single tool to accomplish all of this, some existing tools offer a great deal of functionality, especially in the area of maintaining a single vocabulary.
One tool, Protégé, is particularly well suited to maintaining concepts and relationships within a vocabulary.2 Its support for the creation and maintenance of ontologies, including taxonomies, classifications and their associated vocabulary make it an ideal tool for managing a distinct vocabulary. The master vocabulary, in turn, provides the base hierarchy upon which to map other vocabulary sources. The problem, then, reduces to managing mappings to outside source vocabularies, which itself, is not a small task.
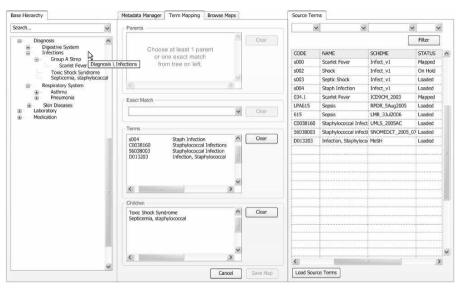
A new mapping tool shown above will integrate with Protégé and facilitate mappings between the base hierarchy and source terms. This tool allows a user to efficiently locate source and base concepts and create multiple mappings, reducing the time and cost of maintaining vocabulary relationships while enforcing the correct management of an Ontology through Protégé. By leveraging the strengths of Protégé and addressing the mapping problem with a new tool within the i2b2 framework, we can create a powerful solution that will make possible the goal of integrating biology and the bedside.
Acknowledgments
This work was funded by the NIH Roadmap for Medical Research, Grant U54LM008748.
References
- 1.National Information Standards Organization. 2005. Information and documentation - Guidelines for the Construction, Format, and Management of Monolingual Controlled Vocabularies; p. 172. (ANSI/NISO Z39.19-2005) [Google Scholar]
- 2.The Protégé Ontology Editor and Knowledge Acquisition System. [homepage on the Internet] Stanford. Stanford Medical Informatics. c2005–2006 [cited 2006 March 13]. Available from: http://protege.stanford.edu/