Abstract
Integrative neuroscience involves the integration and analysis of diverse types of neuroscience data involving many different experimental techniques. This data will increasingly be distributed across many heterogeneous databases that are web-accessible. Currently, these databases do not expose their schemas (database structures) and their contents to web applications/agents in a standardized, machine-friendly way. This limits database interoperation. To address this problem, we describe a pilot project that illustrates how neuroscience databases can be expressed using the Web Ontology Language, which is a semantically-rich ontological language, as a common data representation language to facilitate complex cross-database queries. In this pilot project, an existing tool called “D2RQ” was used to translate two neuroscience databases (NeuronDB and CoCoDat) into OWL, and the resulting OWL ontologies were then merged. An OWL-based reasoner (Racer) was then used to provide a sophisticated query language (nRQL) to perform integrated queries across the two databases based on the merged ontology. This pilot project is one step toward exploring the use of semantic web technologies in the neurosciences.
INTRODUCTION
Integrative neuroscience [1–4] involves studying how the brain operates by exploring the intricate interrelationship of its multiple levels of functional organization (starting at the genetic level, moving up through the synaptic, neuronal, and brain-pathway levels, and ultimately reaching the behavioral level). The integration of these different levels of information has the potential to provide the neuroscientist with a better understanding of how the brain functions as well as the mechanisms of many human neurological diseases.
The field of neuroscience has generated a large quantity of data that feature high complexity and diversity. These data have been derived from a wide variety of experimental approaches including high-throughput techniques (e.g., DNA microarrays [5] and Mass Spectrometry [6]), and involve samples derived from different types of tissues and cells (e.g., Purkinje cells and glial cells), under different experimental and clinical conditions, located at different regions of the brain (e.g., cerebral cortex and cerebellum) in different organisms (e.g., mouse and human).
In addition to data diversity, a major obstacle to integration of neuroscience data has been the proliferation of machine-unfriendly formats used by different data providers to expose their data. Since many of the current neuroscience databases are web-accessible, the HyperText Markup Language (HTML) has been a popular format for presenting neuroscience data to the human user. Other formats (which are also frequently used for web data display and export) include tab-delimited format, binary (image) format, and free text. While these formats are human-friendly for data display purposes, they lack explicit semantic (ontological) description of the data, which is required for “intelligent” processing by machine. Using these formats, more efforts are needed to parse the data and to capture the data semantics in the code logic. Not only does this make data integration difficult, but it also creates a significant software maintenance problem when the format and/or meaning of the data change. It should be straightforward to translate from a machine-friendly format to a human-friendly format (but not vice versa).
Traditional approaches to database integration including data federation (e.g., QIS [7]) and data warehouse (e.g., BioWarehouse [8]) involve mapping between the component data models (e.g., relational data model) and a common data model (e.g., object-oriented data model). To go beyond a data model, the semantic web approach relies on using an ontology to integrate different databases. Unlike data models which concern about how to use data structures to convey meanings, ontologies are concerned about the meanings being conveyed. In addition, the fundamental asset of ontologies is their relative independence of particular applications. That is, an ontology consists of relatively generic knowledge that can be reused by different kinds of applications. Several ontological languages (implemented based on the eXtensible Markup Language or XML) have been developed to help expose ontologies widely over the web.
Our approach involves developing a standardized machine-friendly data representation to expose neuroscience data to web applications/agents that perform data integration and analysis. To this end, we are exploring the use of the Web Ontology Language (OWL) (http://www.w3.org/TR/owl-features/), a computationally expressive ontological language, to encode neuroscience data. OWL represents a step in the direction of the semantic web (next generation of the web) [9] where data are self-described and meaningful to computers through ontological languages ranging from the Resource Description Framework (RDF) (http://www.w3.org/RDF/), RDF Schema (RDFS) (http://www.w3.org/TR/rdf-schema/), and OWL with increasing expressive power. The goal of the semantic web is to enable computers to do a better job in helping human users in discovering knowledge based on web-accessible data. Current approaches to neuroscience database integration (e.g.,[1, 7, 10]) have not extensively explored the emerging semantic web technology, which has received a growing attention in the life sciences community [11–13]. In addition, biological datasets such as UniProt [14] and Gene Ontology [15] have recently made available in RDF format. The contribution of this paper lies in exploring the use of a semantically-expressive ontological language in database interoperation in the neuroscience domain. Such a language provides a powerful inferencing capability and a construct that allows semantic correspondences to be established between different ontologies.
METHOD
To demonstrate the potential benefits of the semantic web in the neurosciences, we provide a pilot use case that shows how several semantic web tools can be used to facilitate integration of two neuroscience databases: NeuronDB [16] and CoCoDat [17].
NeuronDB (http://senselab.med.yale.edu/senselab/NeuronDB/), which is a sub-database of SenseLab [18], captures data that characterize three types of neuronal membrane properties (1) ionic channels, 2) neuron-transmitter receptors, and 3) neuro-transmitter substances) in different compartments (e.g., soma, distal apical dendrite) of different neurons (e.g., Mitral cell, Purkinje cell) in different regions of the brain (e.g., neocortex, cerebellum, and hippocampus). It provides a number of web-based search tools that allow for integration of these properties in a given type of neuron and compartment, and for comparison of properties across different types of neurons and compartments. NeuronDB (and SenseLab as a whole) is implemented using Oracle with a web interface.
CoCoDat (http://www.cocomac.org/cocodat/) is a neuronal microcircuitry database that contains bibliographic references as well as data and parameter values from published experimental reports. The data contained in CoCoDat characterize the experimental procedures, the brain structure (region, layer, neuron type and cellular compartment) with a focus on the neo-cortex, as well as the experimental results obtained in the six categories: 1) morphology, 2) firing properties, 3) ionic currents, 4) ionic conductances, 5) synaptic currents, and 6) connectivity. In addition to providing a web interface for viewing the catalog presentation of the data (without searching capabilities), CoCoDat is available for download as a Microsoft Access 2000 database (with a search interface) running on local computer.
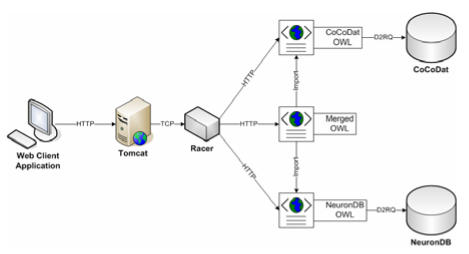
Figure 1 depicts the components of our prototype system that serves as a pilot demonstration. The target OWL structure of each database is designed manually using Protégé (http://protege.stanford.edu/), an ontology editor with OWL support. Then we use a tool called D2RQ (http://www.wiwiss.fuberlin.de/suhl/bizer/D2RQ/), which provides a high-level mapping specification language to specify rules for translating a relational database into the target RDF or OWL ontology, to automatically translate the two neuroscience databases (NeuronDB and CoCoDat) into their corresponding OWL ontological structures. The translation is based on the mapping rules created manually according to the original database schema and their corresponding OWL structures. Next we import the two translated ontologies into a single ontology and semantically link the ontological terms using OWL. Such linking or merging often requires neuroscience expertise to identify related concepts in the two different ontologies. Finally we use an OWL-based reasoner (Racer [19]) that provides a sophisticated query language called nRQL to retrieve and integrate data from NeuronDB and CoCoDat based on the merged ontology. A web interface (using Tomcat) is then created for users to download the individual OWL ontologies (with or without data), the merged ontology, and the D2RQ mapping rules. The web interface also allows the user to compose nRQL queries and displays the query results.
Figure 1.
System architecture.
RESULTS
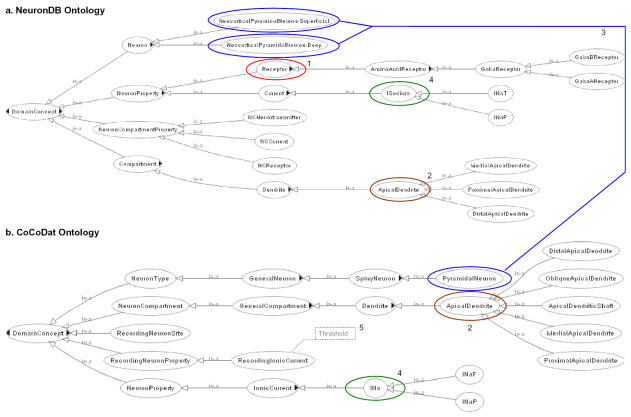
Figure 2 (a) shows graphically a portion of the OWL ontology corresponding to NeuronDB and (b) a portion of the OWL ontology corresponding to CoCoDat. To simplify the graphical representation, the class properties are not shown. As shown in the figure, the ontologies contain synonymous concepts. For example, the concept “ionic sodium current” is represented by the class “ISodium” in NeuronDB, whereas the same concept is represented by the class “INa” in CoCoDat. The figure also indicates a connection between the two ontologies. This connection represents the “SameAs” construct in OWL, which allows one to relate equivalent concepts between different ontologies. In this example, the concept “pyramidal neuron” in CoCoDat is specified to be the same as “neocortical pyramidal neuron: superficial” or “neocortical pyramidal neuron: deep” in NeuronDB. This example also illustrates that different types of neurons are modeled using different hierarchical structures in NeuronDB and CoCoDat. In the case of NeuronDB, different types of neurons are modeled as subclasses (e.g., “neocortical pyramidal neuron: superficial” and “neocortical pyramidal neuron: deep”) of neuron. Each of these subclasses captures information about a region, layer and neuron type. In the case of CoCoDat, some of these components are modeled by separate classes (e.g., neuron type and layer). CoCoDat does not explicitly provide information about the brain region, as it is implicit in the fact that CoCoDat is only concerned with the neocortex region.
Figure 2.
(a) A portion of the NeuronDB ontology and (b) a portion of the CoCoDat ontology.
Figure 3 shows an nRQL statement that queries both NeuronDB and CoCoDat in an integrated fashion. The query does the following:
Figure 3.
A cross-database query written in nRQL syntax and the corresponding answer.
It retrieves the receptors1 contained in the apical dendrite2compartment of all types of pyramidal neurons3 in the neocortex, which have been measured for an ionic sodium4current having a voltage “threshold” 5 of at least −35 mV.
The underlined terms and superscripts correspond to concepts defined in the ontologies (these concepts are circled in the ontology diagram shown in Fig. 2). This query illustrates how NeuronDB and CoCoDat are complementary to each other. In this query example, receptors are contained only in NeuronDB, while the threshold for ionic sodium current is available only in CoCoDat. It also demonstrates the inferencing capability supported by OWL. For example, although the query only specifies “apical dendrite” as the compartment, it automatically traverses its subclass hierarchy to retrieve data from all of its child classes including “distal apical dendrite” and “oblique apical dendrite” (as shown in Figure 2b). This integrated query is made possible by the “SameAs” relationship defined among different types of pyramidal neurons between the NeuronDB and CoCoDat ontologies. Using this OWL construct, we can retrieve data across different levels of granularity. In this case, the query returns receptors of pyramidal neurons across different layers of the neocortical region. The results of the query are shown in Figure 3. As shown in the figure, four receptors are found.
DISCUSSION
We demonstrated how to use the OWL “SameAs” construct to merge two different neuroscience ontologies converted from two existing neuroscience databases – CoCoDat and NeuroDB. In our example, we merged the ontologies by the “pyramidal neuron” concept. We showed that the expressivity of OWL enables us to define seemingly different neuron types as identical and to inference the data with the added definition. We observed that the different categorization of neuron types in different databases may lead to a more complicated integration involving mapping a single concept in one ontology to a set of related concepts in another ontology.
As the number of web-accessible neuroscience resources (including databases and tools) continues to increase, it will become more difficult for users to find these resources, decide which ones are potentially relevant to their research studies, and even integrate the resources to meet their needs. The more the number of the resources we integrate, the higher the complexity the integration would be. However, if the ontologies of the web resources can be created on top of those upper standard ontologies such as the Functional Genomics Investigation Ontology (FuGO) (http://fugo.sourceforge.net/), integration of an increasing number of neuroscience databases would not be exponentially complex. Moreover, keeping track of the ontology versions may also ease the maintainability of the ontology while the content changes over time. For instance, the “priorVersion”, “backwardCompatibleWith” and “incompatibleWith” constructs in OWL could be used to keep track and control the versions of the entire ontology while the “DeprecatedClass” and “DeprecatedProperty" constructs could be used to maintain version information for classes and properties.
Our current pilot demonstration is based on the assumption that a user knows exactly the location of the databases s/he wants to integrate. With the current web technology, many users rely on search engines such as Google to help them find the resources they want through keyword searches. This approach is problematic in terms of sensitivity and specificity. For example, the user may need to sift through a large number of hits returned by a search engine and still may not find what he wants. As an alternative approach, the user can visit a central registry such as Neuroscience Database Gateway (http://apu.sfn.org/content/Programs/NeuroscienceDatabaseGateway) and neuroguide.com (http://www.neuroguide.com), which lists the resources registered by their providers or compiled by a single person/group. Such a registry typically provides some categorical organization and structured search interface to help the user find the resources s/he needs.
Despite their usefulness, these registries are limited by a lack of standards since different registries use different vocabularies or ontologies to describe their entries. In addition, they do not provide a detailed enough database description or inferencing capability to allow the user to retrieve data in an integrated flexible fashion. Semantic web technologies may help point the way to solutions to this problem. For example, if the developers of CoCoDat and NeuronDB agree to expose their database description as OWL ontologies, we could take advantage of the detailed ontological description and the inferencing feature supported by OWL to perform more intelligent searches. For example, if a user enters the term “pyramidal neuron”, the search engine would not only return CoCoDat as the only resource (even though the explicit “pyramidal neuron” concept only occurs in the CoCoDat ontology), but it would also return NeuronDB as the additional relevant resource (because the NeuronDB ontology contains concepts “neocortical pyramidal neuron: superficial” and “neocortical pyramidal neuron deep” which are specified to be the same as the “pyramidal neuron” concept defined in the CoCoDat ontology). Swoogle [20] is an example of a semantic-web-based search engine that allows registration and searching of RDF or OWL ontologies over the web.
While advanced ontological languages such as OWL provide expressive data modeling constructs, their querying and reasoning capability is limited by the size of the ontology (and the associated data) involved. In our demonstration, the Racer reasoner that we used takes 8 seconds to execute the query in Figure 3 with about 1000 records on a Pentium 4 1.69 GHz machine with 512 MB RAM. There is clearly a tradeoff between expressivity and scalability. There are OWL reasoners such as OWLIM (http://www.ontotext.com/owlim/) that can handle large ontologies, but they sacrifice the support of expressive queries that might be needed in some cases. How to increase the performance of large-scale and complex data reasoning is an active area of computer science research. In the case of neuroscience data integration and reasoning, we need to develop use cases to see how to strike a sensible balance between expressivity and performance. For very large neuroscience datasets, we may consider using less expressive languages such as RDF to encode the ontologies, as they can be handled more efficiently by current semantic web technologies. Our current pilot project is one step toward exploring the use of semantic web technologies in the neurosciences.
CONCLUSION
We have presented a pilot semantic web use case in the neuroscience domain. Specifically, we have demonstrated how to use OWL as a common language for representing the ontologies and their associated data corresponding to different neuroscience databases. We also showed how these ontologies can be merged using OWL as well as queried by nRQL using Racer. While there are limitations in the current semantic web technologies, our prototype use case represents an example illustrating how integrative neuroscience and semantic web can intersect.
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants K25 HG02378, P01 DC04732, T15 LM 07056, P20 LM07253, and NSF grant DBI-0135442.
REFERENCES
- 1.Martone ME, Gupta A, Ellisman MH. e-Neuroscience: challenges and triumphs in integrating distributed data from molecules to brains. Nat Biotechnol. 2004;7(5):467–72. doi: 10.1038/nn1229. [DOI] [PubMed] [Google Scholar]
- 2.Miller G. Neuroscience. New neurons strive to fit in. Science. 2006;311(5763):938–40. doi: 10.1126/science.311.5763.938. [DOI] [PubMed] [Google Scholar]
- 3.Koslow SH. Discovery and integrative neuroscience. Clin EEG Neurosci. 2005;36(2):55–63. doi: 10.1177/155005940503600204. [DOI] [PubMed] [Google Scholar]
- 4.Gordon E. Integrative neuroscience. Neuropsychopharmacology. 2003;28(Suppl):2–8. doi: 10.1038/sj.npp.1300136. [DOI] [PubMed] [Google Scholar]
- 5.Morris CM, Wilson KE. High throughput approaches in neuroscience. Int J Dev Neurosci. 2004;22(7):515–22. doi: 10.1016/j.ijdevneu.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Husi H, Grant SG. Proteomics of the nervous system. Trends Neurosci. 2001;24:259–66. doi: 10.1016/s0166-2236(00)01792-6. [DOI] [PubMed] [Google Scholar]
- 7.Marenco L, Wang TY, Shepherd G, Miller PL, Nadkarni P. QIS: A framework for biomedical database federation. J Am Med Inform Assoc. 2004;11(6):523–34. doi: 10.1197/jamia.M1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.T.J. Lee, Y. Pouliot, V. Wagner, P. Gupta, D.W. Stringer-Calvert, J.D. Tenenbaum, P.D. Karp2006BioWarehouse: a bioinformatics database warehouse toolkit Bioinformatics 7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berners-Lee T, Hendler J, Lassila O.2001The semantic web Scientific American 284534–43.11396337 [Google Scholar]
- 10.Gardner D, Knuth KH, Abato M, Erde SM, White T, DeBellis R, Gardner EP. Common data model for neuroscience data and data model exchange. J Am Med Inform Assoc. 2001;8(1):17–33. doi: 10.1136/jamia.2001.0080017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung K-H, Yip KY, Smith A, deKnikker R, Masiar A, Gerstein M. YeastHub: a semantic web use case for integrating data in the life sciences domain. Bioinformatics. 2005;21(suppl_1):i85–96. doi: 10.1093/bioinformatics/bti1026. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Gorlitsky R, Almeida JS. From XML to RDF: how semantic web technologies will change the design of 'omic' standards. Nat Biotechnol. 2005;23(9):1099–103. doi: 10.1038/nbt1139. [DOI] [PubMed] [Google Scholar]
- 13.Neumann EK, Quan D. Biodash: A Semantic Web Dashboard for Drug Development. Pacific Symposium on Biocomputing. World Scientific; 2006. pp. 176–87. [PubMed] [Google Scholar]
- 14.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucl Acids Res. 2004;32(90001):D115–119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry M, Davis A, Dolinski K, Dwight S, Eppig J, et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirsky JS, Nadkarni PM, Healy MD, Miller PL, Shepherd GM. Database tools for integrating and searching membrane property data correlated with neuronal morphology. J Neurosci Methods. 1998;82(1):105–21. doi: 10.1016/s0165-0270(98)00049-1. [DOI] [PubMed] [Google Scholar]
- 17.Dyhrfjeld-Johnsen J, Maier J, Schubert D, Staiger J, Luhmann HJ, Stephan KE, Kotter R. CoCoDat: a database system for organizing and selecting quantitative data on single neurons and neuronal microcircuitry. J Neurosci Methods. 2005;141(2):291–308. doi: 10.1016/j.jneumeth.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Skoufos E, Marenco L, Nadkarni PM, Miller PL, Shepherd GM. Olfactory receptor database: a sensory chemoreceptor resource. Nucl Acids Res. 2002;28(1):341–3. doi: 10.1093/nar/28.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haarslev V, Möller R, Straeten RVD, Wessel M. 2004 International Workshop on Description Logics (DL-2004) 2004. Extended Query Facilities for Racer and an Application to Software-Engineering Problems; pp. 148–57. [Google Scholar]
- 20.Li D, Finn T, Joshi A, Pan R, Cost RS, Peng Y, Reddivari P, Doshi VC, Sachs J. Proceedings of the Thirteenth ACM Conference on Information and Knowledge Management. ACM Press; 2004. Swoogle: A Search and Metadata Engine for the Semantic Web; pp. 652–9. [Google Scholar]