Abstract
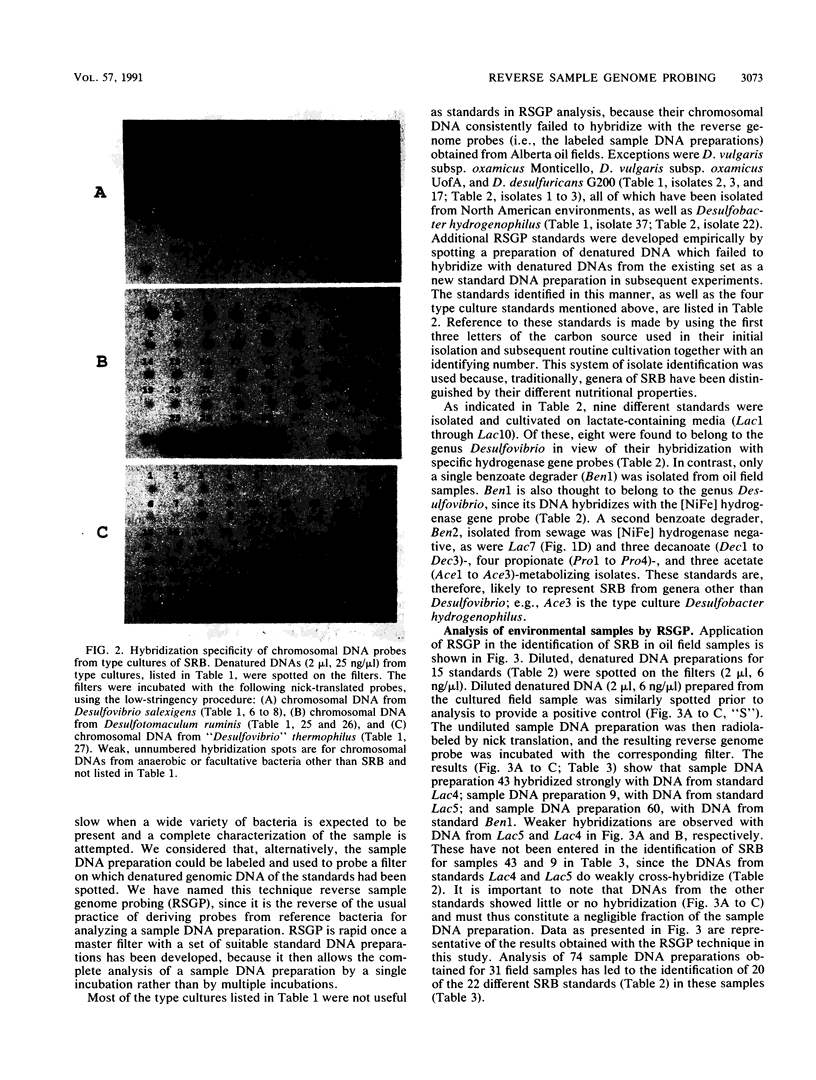
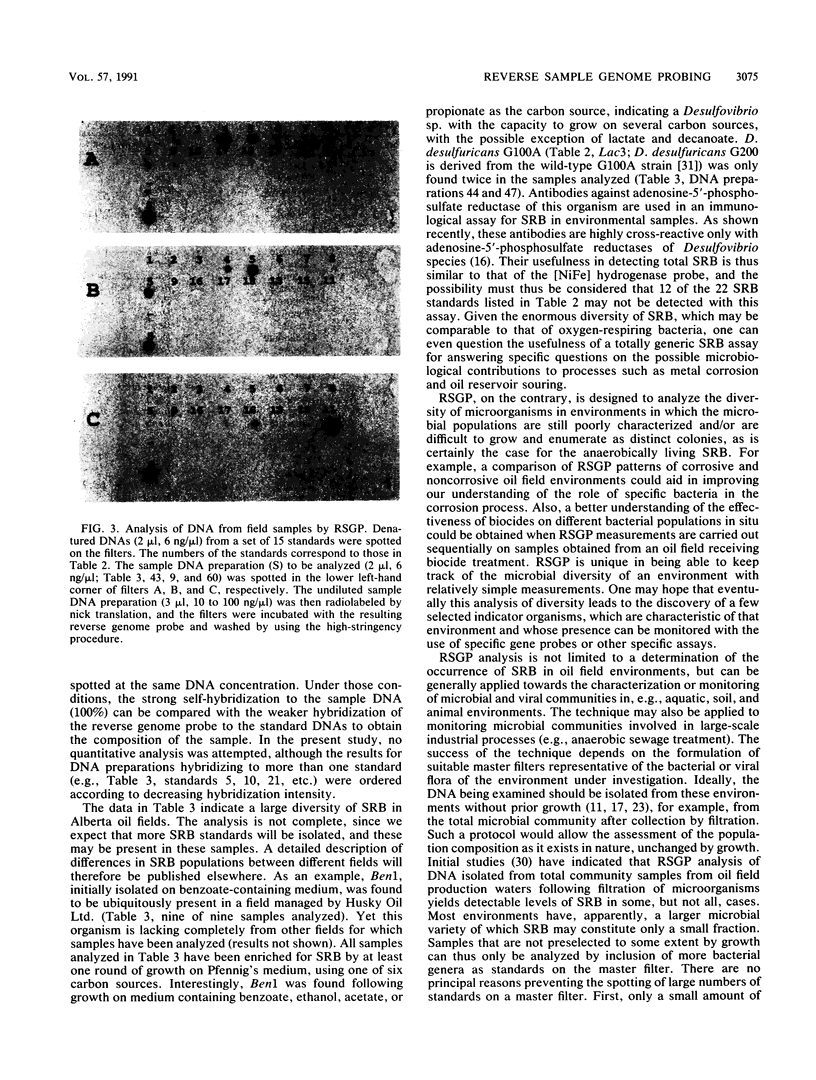
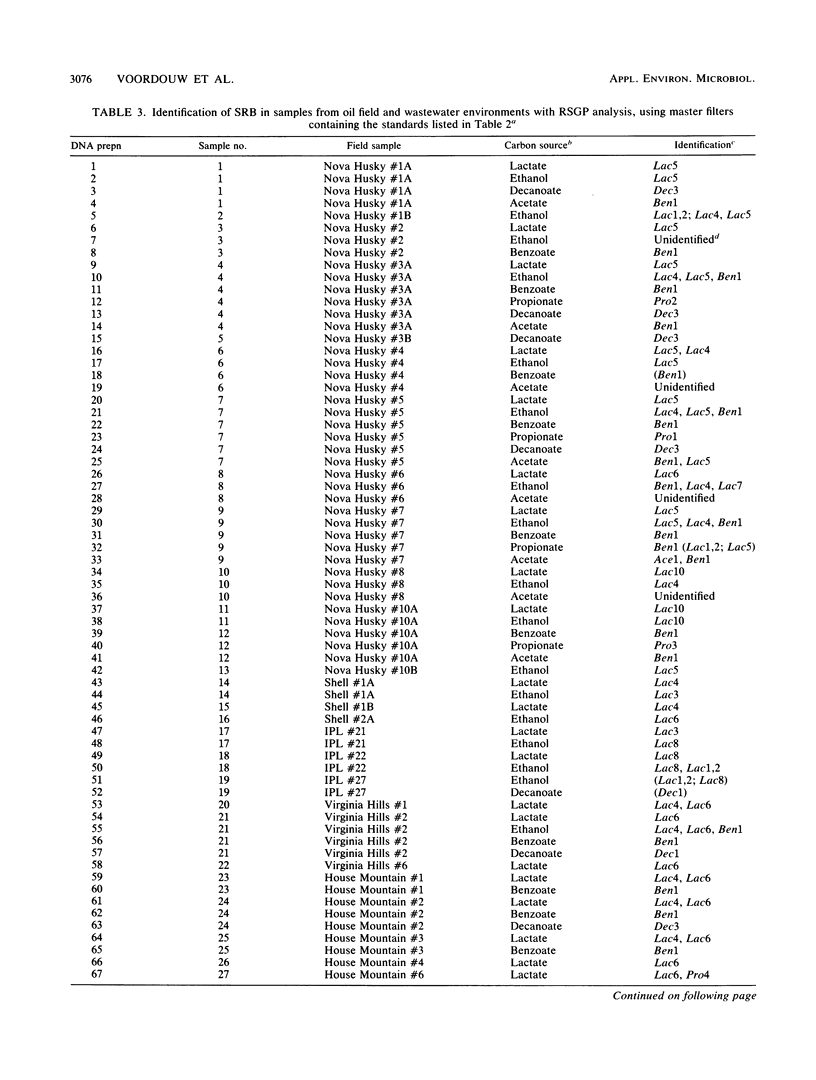
A novel method for the identification of bacteria in environmental samples by DNA hybridization is presented. It is based on the fact that, even within a genus, the genomes of different bacteria may have little overall sequence homology. This allows the use of the labeled genomic DNA of a given bacterium (referred to as a “standard”) to probe for its presence and that of bacteria with highly homologous genomes in total DNA obtained from an environmental sample. Alternatively, total DNA extracted from the sample can be labeled and used to probe filters on which denatured chromosomal DNA from relevant bacterial standards has been spotted. The latter technique is referred to as reverse sample genome probing, since it is the reverse of the usual practice of deriving probes from reference bacteria for analyzing a DNA sample. Reverse sample genome probing allows identification of bacteria in a sample in a single step once a master filter with suitable standards has been developed. Application of reverse sample genome probing to the identification of sulfate-reducing bacteria in 31 samples obtained primarily from oil fields in the province of Alberta has indicated that there are at least 20 genotypically different sulfate-reducing bacteria in these samples.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkay T., Liebert C., Gillman M. Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants: mer genes and Hg2+ resistance. Appl Environ Microbiol. 1989 Jun;55(6):1574–1577. doi: 10.1128/aem.55.6.1574-1577.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry G. R., Toranzos G. A., Bhatti A. R. Novel method for monitoring genetically engineered microorganisms in the environment. Appl Environ Microbiol. 1989 May;55(5):1301–1304. doi: 10.1128/aem.55.5.1301-1304.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry G. R., Toranzos G. A., Bhatti A. R. Novel method for monitoring genetically engineered microorganisms in the environment. Appl Environ Microbiol. 1989 May;55(5):1301–1304. doi: 10.1128/aem.55.5.1301-1304.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux R., Delaney M., Widdel F., Stahl D. A. Natural relationships among sulfate-reducing eubacteria. J Bacteriol. 1989 Dec;171(12):6689–6695. doi: 10.1128/jb.171.12.6689-6695.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux R., He S. H., Doyle C. L., Orkland S., Stahl D. A., LeGall J., Whitman W. B. Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J Bacteriol. 1990 Jul;172(7):3609–3619. doi: 10.1128/jb.172.7.3609-3619.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diels L., Mergeay M. DNA probe-mediated detection of resistant bacteria from soils highly polluted by heavy metals. Appl Environ Microbiol. 1990 May;56(5):1485–1491. doi: 10.1128/aem.56.5.1485-1491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D., Starrenburg M. J., Akkermans A. D. Oligonucleotide Probes That Hybridize with rRNA as a Tool To Study Frankia Strains in Root Nodules. Appl Environ Microbiol. 1990 May;56(5):1342–1346. doi: 10.1128/aem.56.5.1342-1346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating S. T., Burand J. P., Elkinton J. S. DNA hybridization assay for detection of gypsy moth nuclear polyhedrosis virus in infected gypsy moth (Lymantria dispar L.) larvae. Appl Environ Microbiol. 1989 Nov;55(11):2749–2754. doi: 10.1128/aem.55.11.2749-2754.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotomi M., Ohno T., Mutai M. Rapid and correct identification of intestinal Bacteroides spp. with chromosomal DNA probes by whole-cell dot blot hybridization. Appl Environ Microbiol. 1988 May;54(5):1158–1162. doi: 10.1128/aem.54.5.1158-1162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom J. M., Jessie K., Knodel E., Emptage M. Immunological cross-reactivities of adenosine-5'-phosphosulfate reductases from sulfate-reducing and sulfide-oxidizing bacteria. Appl Environ Microbiol. 1991 Mar;57(3):727–733. doi: 10.1128/aem.57.3.727-733.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Walsh P. S., Levenson C. H., Erlich H. A. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadpour M., Liston J., Ongerth J. E., Tarr P. I. Evaluation of DNA probes for detection of Shiga-like-toxin-producing Escherichia coli in food and calf fecal samples. Appl Environ Microbiol. 1990 May;56(5):1212–1215. doi: 10.1128/aem.56.5.1212-1215.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. C., Knight I. T., Straube W. L., Colwell R. R. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989 Mar;55(3):548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spierings G., Hofstra H., Huis in'T Veld J., Hoekstra W., Tommassen J. Development of enterobacterium-specific oligonucleotide probes based on the surface-exposed regions of outer membrane proteins. Appl Environ Microbiol. 1989 Dec;55(12):3250–3252. doi: 10.1128/aem.55.12.3250-3252.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G. W. Biotin-labeled plasmid DNA probes for detection of epithelium-associated strains of lactobacilli. Appl Environ Microbiol. 1989 Feb;55(2):461–464. doi: 10.1128/aem.55.2.461-464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G., Niviere V., Ferris F. G., Fedorak P. M., Westlake D. W. Distribution of Hydrogenase Genes in Desulfovibrio spp. and Their Use in Identification of Species from the Oil Field Environment. Appl Environ Microbiol. 1990 Dec;56(12):3748–3754. doi: 10.1128/aem.56.12.3748-3754.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G., Strang J. D., Wilson F. R. Organization of the genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J Bacteriol. 1989 Jul;171(7):3881–3889. doi: 10.1128/jb.171.7.3881-3889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer P. J., Van Kavelaar M. J., Michel C. B., Ng T. K. Effect of Phosphate on the Corrosion of Carbon Steel and on the Composition of Corrosion Products in Two-Stage Continuous Cultures of Desulfovibrio desulfuricans. Appl Environ Microbiol. 1988 Feb;54(2):386–396. doi: 10.1128/aem.54.2.386-396.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]