The Informatics for Integrating Biology and the Bedside (i2b2) is one of the sponsored initiatives of the NIH Roadmap National Centers for Biomedical Computing (http://www.bisti.nih.gov/ncbc/). One of the goals of i2b2 is to provide clinical investigators broadly with the software tools necessary to collect and manage project-related clinical research data in the genomics age as a cohesive entity – a software suite to construct and manage the modern clinical research chart.
The i2b2 team is developing an interoperable software framework for the research chart that can be extended for new and unanticipated data types as well as functionality. It is intended to serve the following users:
Clinical investigators who want to use the software in as “shrink-wrapped” a way as possible,
Bioinformatics scientists who want the ability to customize the flow of data and interactions, and
Biocomputational software developers who want to develop new software capabilities that can be integrated easily into the computing environment.
One method for developing this new software and framework is to work hand and hand with current researches to access there current and future needs. Our Asthma “Driving Biology Project” studies the interplay between environmental exposures and genetic variation in determining both individual airways disease risk and individual response to airways disease medications. Tools and methods currently available were successful for monogenic disorders, but have not yielded major breakthroughs in complex diseases to date. This work will lead to the development and implementation of methods and tools to improve genetic epidemiological and pharmacogenetic research in complex disease.
In order to run queries and perform data mining to determine the differential affects of environmental exposures such as smoking on asthma patients, the Asthma DBP created a data mart containing 71 million clinical observations from over 95 thousand patients. Research specific data not routinely available in clinical data sets was loaded by running it through a set of web services. These services could be located within the local network or on a different remote network. Each of the services is either exclusively a web service or a web service wrapper is developed to encumber the native service. Because each of the i2b2 web services, expect and produce the same xml schema, they can be interconnected in multiple different ways.
For example, Concept-Value Pair Extraction from Semi-Structured Clinical Reports1 was wrapped into a web service to process pulmonary reports, which extracted pre and post bronchodilator FEV1, FVC, and patient vital signs. Also using patient notes from discharge summaries and the electronic medical record, a Natural Language Processing service was developed to determine if a patient was a current, past or never a smoker, as well as asthma related medications and diagnoses.
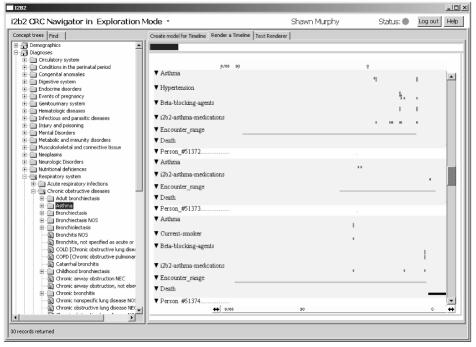
Finally, Query2 and Visualization tools (above) allow the researcher to have a visual representation of the data in a clear and informative viewpoint that allow exploration of the clinical and genetic data.
Acknowledgments
This work was funded by the NIH Roadmap for Medical Research, Grant U54LM008748.
References
- 1.Chung J, Murphy SN. Concept-Value Pair Extraction from Semi-Structured Clinical Narrative: A Case Study Using Echocardiogram Reports. JAMIA Symposium. 2005:131–5. [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy SN, Gainer VS, Chueh H. A Visual Interface Designed for Novice Users to find Research Patient Cohorts in a Large Biomedical Database. JAMIA Symposium. 2003:489–3. [PMC free article] [PubMed] [Google Scholar]