Abstract
Progression of pro-B lymphocytes to the pre-B stage depends on the expression of a pre-B cell receptor (pre-BCR), consisting of an Ig μ H chain, Ig surrogate light chain, and associated signal transducing chains. Mice that are unable to express a pre-BCR show an arrest of B cell development at the pro-B stage. Such is the case for severe combined immune deficient (SCID) mice in which μ chains are not made because of a defect in V(D)J recombination. When μ chains are made, as in SCID mice bearing a functional μ transgene, then B cell differentiation can proceed to the pre-B stage. However, as reported here, a μ transgene (M54) that promotes development of SCID pre-B cells in adult bone marrow fails to do so in fetal liver. We suggest that a pre-BCR containing the M54 μ chain cannot signal progression of pro-B cells to the pre-B stage in the fetal liver microenvironment.
Keywords: B cell differentiation
According to the current paradigm for B lymphocyte development (reviewed in refs. 1–4), differentiation of pro-B cells into pre-B cells requires the expression of a cell surface receptor, termed the pre-B cell receptor (pre-BCR). The pre-BCR contains an Ig μ and Ig surrogate light (SL) chain and associated signal transducing chains. Similarly, the differentiation of pre-B cells into B cells requires the expression of a cell surface BCR, which differs from a pre-BCR in that it contains a conventional Ig L chain instead of a SL chain. In support of this paradigm, B cell development in adult bone marrow is blocked or severely impaired in mice with genetic lesions that prevent expression of a pre-BCR or BCR (5–10). Further, the development of pre-B cells in adult bone marrow has been shown to depend on the expression of μ chains that strongly associate with SL chain (11, 12).
As a pre-BCR cannot be made in mice homozygous for the severe combined immune deficient (SCID) mutation (SCID mice), B cell differentiation in these mutant mice is arrested at the pro-B stage (reviewed in ref. 13), the stage at which H chain gene rearrangement is initiated (14–16). The arrest reflects the inability of developing SCID B lineage cells to rearrange VH, DH, and JH gene elements productively (17). However, if the requirement for VDJH gene rearrangement is circumvented by selectively crossing the μ transgene (tg), 3H9 (18), or M54 (19) into the SCID genome, then a pre-BCR can be expressed and differentiation of bone marrow pro-B cells proceeds to the pre-B stage (5, 20). Differentiation of SCID pre-B cells to the B cell stage requires the presence of a L chain tg, such as Vκ8 (21), in addition to 3H9 or M54. M54/Vκ8 and 3H9/Vκ8 mice can form a BCR and generate B cells in their bone marrow and spleen (20).
Although the above tgs have been shown to promote SCID B cell differentiation in adult bone marrow, it is open to question whether they would have a similar effect on SCID B cell differentiation in fetal liver. Wasserman et al. (22) recently reported that a human μ tg that promotes pre-B cell development in adult bone marrow of recombination activation gene 1 (RAG1)-deficient mice (23), fails to promote significant development of pre-B cells in fetal liver of RAG1-deficient mice. Conversely, in another μ-tg (VH11) line of RAG1-deficient mice with a low tg copy number, the development of pre-B cells was found to be promoted in fetal liver but not adult bone marrow. VH11 μ chains were shown to associate poorly with SL chain (22), implying that the fate of a pro-B cell in a given tissue microenvironment may vary depending on how well its μ chain interacts with SL chain.
The fate of a pro-B cell in fetal liver or adult bone marrow also might vary depending on the specificity of its pre-BCR. As the representation and diversity of expressed VH regions differ in B cells of fetal and adult mice (24–29), the repertoire of pre-BCR (and BCR) specificities at these different stages of development should be distinct. This leaves open the possibility that a pre-BCR with a μ chain derived from the adult BCR repertoire (such as that coded by 3H9 or M54) might display an inappropriate specificity in fetal liver and fail to signal the transition of SCID fetal pro-B cells to the pre-B stage. Consistent with this possibility, we show here that progression of pro-B cells to the pre-B stage is not promoted in fetal liver of M54 SCID mice, though it is promoted in fetal liver of 3H9 SCID mice. This differential effect cannot be attributed to an obvious difference in association of the M54 or 3H9 μ chain with SL chain. Interestingly, in M54/Vκ8 SCID fetal mice, in which the Vκ8 and SL chains would be expected to compete for pairing with the M54 μ chain, B cell differentiation is clearly promoted as evidenced by a normal number of B cells in the fetal liver of these mice. Based on the effects of 3H9, M54, and M54/Vκ8 on SCID fetal B cell differentiation, we suggest that the specificity of a pre-BCR can influence the fate of developing pro-B cells.
Materials and Methods
Mice.
The selective breeding and genotyping of C.B-17 SCID mice (SCID mice) hemizygous for the μ H chain tg, 3H9 (18), M54 (19), or the kappa L chain tg, Vκ8 (21) have been described (5, 20). Before undertaking this study, M54/+ and Vk8/+ SCID mice were selectively intercrossed to obtain mice homozygous for the respective tgs (homozygosity was confirmed by test crossing suspected homozygotes with non-tg SCID mice). Thus, to obtain M54/+ SCID mice, we crossed M54/M54 SCID mice with non-tg SCID mice. SCID mice bearing both the M54 and Vκ8 tgs (M54/Vκ8 SCID mice) were obtained by crossing M54/M54 and Vκ8/Vκ8 SCID mice. 3H9/+ SCID mice were produced from crosses of 3H9/+ SCID mice with non-tg SCID mice or from intercrosses of 3H9/+ SCID mice; tg+ offspring were identified (subsequent to flow cytometric analysis in the case of embryos) by PCR using tg-specific primers (20). Timed matings were set up over a 12-hr interval (6 p.m. to 6 a.m.), and embryos were analyzed at day 17, 18, or 19. Detection of vaginal plugs confirmed mating (day 0). Adult mice used in this study were between 11 and 22 weeks of age. As a matter of convenience, mice bearing the above tgs are designated with the prefix M54, 3H9, or M54/Vκ8.
Cells.
Single cell suspensions of liver from 17-, 18-, or 19-day embryos were prepared in PBS containing 3% BSA. Cells were treated with 0.165 M NH4Cl to eliminate erythrocytes, washed once with staining medium (RPMI 1640 without phenol red, 10 mM Hepes, 3% FCS, and 0.1% NaN3), resuspended in staining medium and passed through a Nitex sterile nylon screen (Tetko, Elmsford, NY) to remove debris. Bone marrow cells were flushed from femurs of adult mice with staining medium using a syringe and 22-gauge needle. The cells then were dispersed by gentle pipetting, treated with 0.165 M NH4CL, washed and resuspended in staining medium and passed through a sterile nylon screen.
Flow Cytometric Analysis.
Cell suspensions were analyzed for the presence of B lineage cells representing different stages of differentiation as described (20). Classification of pro-B, pre-B, and B cell populations was on the basis of cell surface phenotype alone by using the markers used by Hardy et al. (14). Briefly, cell suspensions were stained with anti-CD45(B220), conjugated with Cy5 (Biological Detection Systems) or allophycocyanin (PharMingen), FITC-conjugated anti-CD43, biotinylated anti-Mac-1, or biotinylated anti-IgM. Binding of biotinylated antibodies was revealed by Texas red-conjugated streptavidin (Southern Biotechnology Associates) or phycoerythrin-conjugated streptavidin (kindly supplied by R. Hardy, Fox Chase Cancer Center, Philadelphia). Pro-B (B220+CD43+IgM−), pre-B (B220+CD43−IgM−), and B (B220+CD43−IgM+) cell populations were enumerated or sorted by multiparameter flow cytometry using a dual laser FACStar Plus (Becton Dickinson). It should be noted that the B220+CD43+IgM− cell population includes several distinct pro-B subsets as well as some non-B lineage cells (30). We subsequently refer to this cell population as the pro-B cell fraction. One experiment included the sorting of an early hematopoietic cell population (B220−CD43+Mac1−) similar to that described by Cumano et al. (31). This cell population is capable of giving rise in culture to monocytes and B cells (Y.C., unpublished results). We refer to this population as pro-M/B cells.
Assay for Expression and Association of μ and SL Chains.
In one series of experiments, the pro-B cell fractions from M54, 3H9, and non-tg SCID fetal liver (day 18) were tested for intracellular expression and association of μ and SL chains. This was done in the manner previously described by using a rat mAb (SL-156) specific for μ-SL chain complexes (32). To obtain sufficient cells for sorting, fetal liver cells from four or more embryos were pooled. To distinguish 3H9 embryos from their non-tg littermates, small aliquots of each embryo cell suspension first were analyzed for the presence of pre-B (B220+CD43−) cells. Cell suspensions containing detectable pre-B cells (i.e., presumptive tg+ embryos, see Fig. 1) then were pooled and sorted for B220+CD43+ cells. The genotype of each embryo donor subsequently was confirmed by PCR using tg-specific primers.
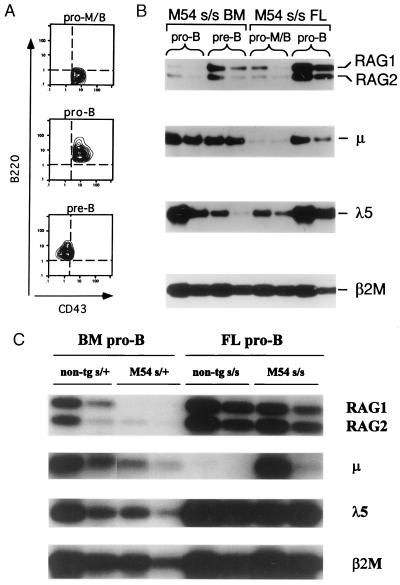
Figure 1.
(A) Development of pre-B cells in 3H9 but not M54 SCID (s/s) fetal mice. Flow cytometric contour plots show B220 versus CD43 staining on IgM− gated cells of day 17 and 19 fetal liver (FL) and adult bone marrow (BM) cells from mice of the indicated genotypes. The numbers within the boxed areas indicate the percentage pre-B (B220+CD43−) cells for the total number of cells analyzed. (B) Development of pre-B cells in γ-irradiated SCID mice injected with M54 or 3H9 fetal liver cells. Adult s/s mice were exposed to 400 cGy and then i.v. injected with 5 × 106 fetal liver cells from day 17 non-tg s/s, 3H9 s/s, M54 s/s, or non-tg s/+ donors. Two to three recipients in each group were analyzed as in A for the presence of bone marrow pre-B cells at 3 and 4 weeks after cell transfer. Representative results are shown for 4 weeks after cell transfer.
Reverse Transcriptase–PCR (RT-PCR).
RNA was obtained from sorted cell subsets by using the method of Chirgwin et al. (33) and reverse-transcribed into cDNA with random oligonucleotides. cDNA corresponding to RAG1, RAG2, μ, the λ5 subunit of the SL chain, and β2-microglobulin (β2M) was PCR amplified for 26 cycles by using oligonucleotides and conditions described previously (34). Amplification of β2M served as an internal control for input cDNA. Blots of RT-PCR-amplified products were hybridized with (i) pCμ3741 (35); (ii), a RAG probe prepared by PCR amplification of RAG1 and RAG2 constructs (provided by D. Schatz, Yale Univ., New Haven, CT); (iii) a λ5 probe prepared by PCR amplification of a λ5 construct (provided by R. Hardy); and (iv), a gel-purified β2M probe made with primers specific for β2M cDNA. Probes were [α-32P]dCTP-labeled by using a Prime-It II kit (Stratagene).
Results
Pre-B Cells Are Detectable in 3H9 But Not M54 SCID Fetal Mice.
To test for the presence of fetal pre-B (B220+CD43−) cells in 3H9 and M54 SCID mice, liver cells from day 17 and day 19 embryos were analyzed by flow cytometry for expression of specific pre-B cell markers. Non-tg SCID and SCID/+ embryos served as negative and positive controls, respectively. Bone marrow cells from SCID, SCID/+, 3H9 SCID, and M54 SCID adult mice also were analyzed. Analyses were performed several times with three or four individual progeny from independent matings. Representative results are shown in Fig. 1A. Note that 3H9 SCID fetal liver contained about 3% pre-B cells (3.3 ± 1.0%) at day 19. Given this result and the approximate 2-fold difference in percentage of pre-B cells in adult bone marrow of 3H9 (10.0 ± 3.6) and M54 (5.3 ± 1.8) SCID mice, we would have expected about 1.5% pre-B cells at day 19 in M54 SCID fetal liver. However, the percentage of pre-B cells in M54 SCID fetal liver was ≤0.3%.
Although M54 SCID fetal liver lacked pre-B cells, i.v. transfer of M54 day 17 fetal liver cells (5 × 106) into γ-irradiated SCID adult recipients resulted in the development of pre-B cells in the host bone marrow (Fig. 1B). At 3 weeks after cell transfer, the percentage of pre-B cells in recipients was about half that in control adult M54 SCID bone marrow, and at 4 weeks, it was equal to the controls (data are shown for the 4-week interval only). Recipients of 3H9 SCID fetal liver cells showed a similar time course for reconstitution of pre-B cells relative to the level of pre-B cells in 3H9 SCID adult bone marrow. Note that pre-B cells were not generated in recipients of fetal liver cells from non-tg SCID donors. We conclude that fetal liver cells from M54 SCID mice are readily able to generate pre-B cells in SCID adult bone marrow.
RAG Expression Is Not Down-Regulated in the Pro-B Cell Fraction of M54 SCID Fetal Mice.
The ability of μ tgs to promote the development of pre-B cells in SCID adult bone marrow has been shown previously to correlate with down-regulated levels of RAG expression in the pro-B (B220+CD43+) cell fraction of these mice (20). Given this correlation and the results of Fig. 1, we suspected that RAG expression might not be down-regulated in the pro-B cell fraction of M54 SCID fetal liver. Without such down-regulation in RAG expression, progression of fetal SCID pro-B cells to the pre-B stage would be impaired because SCID cells cannot efficiently repair RAG-induced DNA double-strand breaks (36). To test for the level of RAG expression, we sorted B220+CD43+ cells from pooled cell suspensions of M54 SCID or non-tg SCID fetal liver (day 17) and compared these sorted cell populations for relative abundance of RAG transcripts. B220+CD43+ cells from non-tg SCID/+ adult bone marrow served as a positive control, and an early hematopoietic cell fraction (B220−CD43+Mac1−) from SCID fetal liver served as a negative control (see Materials and Methods). Contour plots illustrating the phenotype of the different cell subsets analyzed are shown in Fig. 2A.
Figure 2.
RAG expression is down-regulated in the pro-B cell fraction of adult, but not fetal, M54 SCID mice. (A) Flow cytometric contour plots of sorted cells illustrate the different phenotypes of cell populations analyzed: pro-M/B (B220− CD43+IgM−) cells, pro-B (B220+CD43+IgM−), and pre-B (B220+CD43−IgM−) cells. The contour plots shown for pro-M/B and pro-B cells correspond to sorted cells from day 17 fetal liver (FL) of M54 SCID (s/s) mice; the contour plot for pre-B cells corresponds to sorted cells from bone marrow (BM) of M54 s/s adult mice. (B and C) Southern blots of RT-PCR-amplified RAG, μ, and λ5 transcripts in sorted cell populations of adult and day 17 fetal mice. Amplification of the β2M message served as an internal control for input cDNA. The first and second lanes under each bracket (from left to right) correspond to undiluted and 3-fold diluted input cDNA, respectively. The primers and probes used for amplification and hybridization of PCR products are described in Materials and Methods.
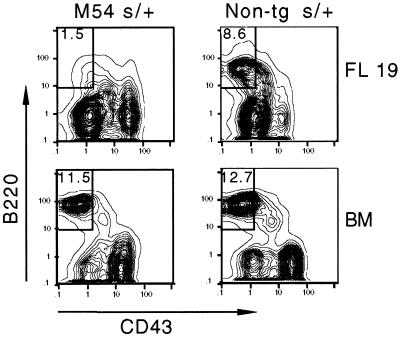
As indicated in Fig. 2 B and C, RAG expression was down-regulated only slightly in the pro-B cell fraction of M54 SCID fetal liver. The levels of RAG1 expression in fetal liver and adult bone marrow of M54 SCID mice relative to the reference control (SCID/+ adult bone marrow) are compared in Table 1. Note that RAG1 expression in M54 SCID fetal mice was comparable to the control and only about 3-fold less than in non-tg SCID fetal mice. Although we would expect uninhibited RAG expression to result in persisting DNA double-strand breaks at DH and JH SCID loci, this may not be the (only) explanation for the lack of pre-B cells in M54 SCID fetal mice. As illustrated in Fig. 3, in M54 SCID/+ fetal mice (heterozygous for the SCID mutation), in which V(D)J recombination is not impaired, the percentage of pre-B cells was about 5-fold less than in the non-tg SCID/+ controls. In contrast, in M54 SCID/+ adult bone marrow, the percentage of pre-B cells was comparable to that in the non-tg SCID/+ controls, consistent with our earlier results (20). These results indicate that M54 has a negative effect on fetal, but not adult, pre-B cell development.
Table 1.
Effect of M54 (and M54/Vκ8) on RAG1, μ, and λ5 expression in pro-B cells of fetal liver and adult bone marrow
| Tg | Genotype | Tissue | RAG1* | μ* | λ5* |
|---|---|---|---|---|---|
| — | s/+ | BM | 1.0 | 1.0 | 1.0 |
| M54 | s/+ | BM | 0.01 | 0.34 | 0.91 |
| M54 | s/s | BM | 0.03 | 0.90 | 1.1 |
| — | s/s | FL | 2.5 | <0.01 | 4.0 |
| M54 | s/s | FL | 0.82 | 1.1 | 1.7 |
| M54/Vκ8 | s/s | FL | 0.11 | 0.23 | 1.1 |
Stored pro-B cells were from adult bone marrow (BM) of SCID/+ (s/+), M54 s/+, and M54 SCID (s/s) mice and from day 17 fetal liver (FL) of s/s, and M54/Vκ8 s/s mice.
The amount of 32P-labeled probe hybridizing to each RT-PCR product (RAG1), λ5, μ, and the internal control (β2M) was quantitated by using a Bio-Image Analyzer. The values shown correspond to the relative amounts of hybridizing signal for RAG1, λ5, and μ each normalized against the internal control (β2M) and the reference control (pro-B cells of s/+ adult bone marrow). Thus for example, the value 0.82 for M54 SCID fetal pro-B cells equals the ratio of the amount of probe hybridizing to RAG1/β2M divided by the amount of probe hybridizing to RAG1/β2M in the reference control.
Figure 3.
Pre-B cell development is impaired in M54 SCID/+ fetal mice. Flow cytometric contour plots show B220 versus CD43 staining on IgM− gated cells from day 19 fetal liver (FL) and adult bone marrow (BM) of M54 SCID/+ (s/+) and non-tg s/+ mice. FL and BM from 4–5 mice of each genotype were analyzed. The results shown are representative. The percentage of pre-B (B220+CD43−) cells is indicated within the boxed areas.
Detection of μ-SL Chain Complexes in M54 and 3H9 SCID Fetal Liver.
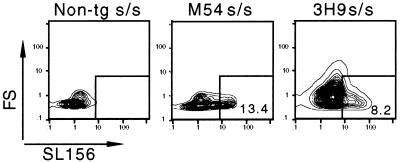
We asked whether the inability of M54 to promote SCID fetal pre-B cell development might reflect poor expression of M54 or poor association of the M54 μ chain with SL chain in fetal pro-B cells. Therefore, we compared the level of μ transcripts in pro-B cell fractions from M54 SCID fetal liver, M54 SCID adult bone marrow and non-tg SCID/+ adult bone marrow. As indicated in Fig. 2 and Table 1, the relative abundance of μ transcripts in each of these cell fractions was comparable, indicating that there was no problem with expression of M54 in SCID fetal liver. Also, the abundance of λ5 transcripts in each of these cell fractions was comparable (λ5 corresponds to a gene coding for a subunit of the SL chain). To test directly for the presence and association of μ and SL chains in M54 SCID fetal mice, we stained the pro-B cell fraction from day 18 fetal liver with an antibody (SL-156) specific for μ-SL chain complexes (32). As shown in Fig. 4, SL-156+ cells were clearly detectable in the pro-B cell fraction from M54 SCID fetal liver. SL-156+ cells also were detectable in the day 18 pro-B cell fraction from 3H9 SCID fetal liver. Thus, we cannot attribute the presence and absence of pre-B cells in 3H9 and M54 SCID fetal mice, respectively, to a gross difference in either expression of the M54 and 3H9 μ chains or association of these μ chains with SL chain.
Figure 4.
Detection of intracellular μ-SL chain complexes in fetal liver of M54 and 3H9 SCID mice. Sorted B220+CD43+IgM− cells from day 18 fetal liver of M54, 3H9, or non-tg SCID embryos were sequentially incubated with biotinylated monoclonal rat SL-156 antibody (or biotinylated rat IgG as a nonspecific control) and phycoerythrin-conjugated streptavidin. Non-tg SCID embryos served as a negative control. Contour plots show forward scatter (FS) versus intensity of intracellular staining with SL-156 antibody. The percentage of SL-156+ cells is indicated within the boxed areas.
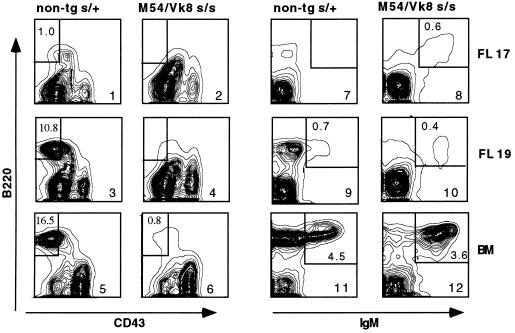
Promotion of B Cell Development in M54/Vκ8 SCID Fetal Mice.
The preceding results suggest that a pre-BCR containing a M54 μ and SL chain is unable to promote SCID fetal B cell differentiation. Therefore, it was of interest to know whether expression of a BCR containing a M54 μ chain and conventional L chain (Vκ8), as in M54/Vκ8 SCID mice, would promote SCID fetal B cell differentiation. In these double-tg mice, Vκ8 chains would be expected to compete with SL chains for pairing with the M54 μ chain. As illustrated in Fig. 5, we found that B cell differentiation indeed is promoted in M54/Vκ8 SCID fetal mice. M54/Vκ8 SCID fetal liver showed a normal percentage (0.4–0.6%) of surface IgM+ B cells (37), which were detectable at day 17, 2 days earlier than in non-tg SCID/+ fetal liver (Fig. 5). As with M54 SCID adult bone marrow, the ability of M54/Vκ8 SCID fetal liver to generate B cells correlated with a marked reduction of RAG expression in the pro-B cell fraction (data are summarized in Table 1). These results suggest that association of M54 μ and Vκ8 chains, as opposed to M54 μ and SL chains, results in a receptor that can promote SCID fetal B cell differentiation. Note that M54/Vκ8 SCID fetal liver and adult bone marrow lacked pre-B cells (Fig. 5, compare panels 3 versus 4 and 5 versus 6). The lack of pre-B cells in M54/Vκ8 SCID mice, despite near-normal numbers of B cells, has been noted earlier (20, 38) and interpreted to reflect rapid or direct progression of tg-expressing pro-B cells to the B cell stage.
Figure 5.
Development of B cells in fetal liver (FL) of M54/Vκ8 SCID embryos. Flow cytometric contour plots on the left show B220 versus CD43 staining on IgM− gated cells and, on the right, B220 versus IgM staining on CD43− gated cells. The percentage of pre-B (B220+CD43−) or B (B220+IgM+) cells is indicated in the boxed areas.
Discussion
μ-tg SCID Mice.
In M54 SCID mice, all developing pro-B cells would be expected to express a pre-BCR, consisting of a M54 μ and SL chain and associated signal transducing chains. We will refer to this receptor as M54/SL. As the progression of bone marrow pro-B cells to the pre-B stage is known to depend on pre-BCR expression (5–12), we assume that the observed pre-B cell development in M54 SCID bone marrow depends on the expression of M54/SL. In M54 SCID fetal liver, however, pre-B cell development was not detectable despite the presence of μ and λ5 transcripts and μ-SL chain complexes in the pro-B cell fraction of this tissue. Thus, we infer that M54/SL is expressed but that it cannot signal progression of fetal pro-B cells to the pre-B stage, consistent with its apparent inability to cause down-regulation of RAG expression in the pro-B cell fraction of fetal SCID liver. The lack of pre-B cell development does not reflect an intrinsic defect of B lineage precursor cells in M54 SCID fetal liver because injection of cells from this tissue into γ-irradiated SCID recipients resulted in the generation of pre-B cells in the host bone marrow.
Why is M54/SL unable to signal the transition of fetal pro-B cells to the pre-B stage? Possibly, M54/SL is not transported to the surface membrane of fetal pro-B cells or it fails to interact with a fetal self-ligand. Either outcome might result in no pre-BCR signaling. Alternatively, although one may assume that each pre-BCR is potentially able to interact with ligand, a pre-BCR ligand interaction may not be essential for the transition of pro-B cells to the pre-B stage (39, 40). Rather, most chance pre-BCR ligand interactions could result in cell deletion, analogous to the deletion of developing B cells that express BCRs with inappropriate anti-self specificities (41–45). Accordingly, one could argue that M54/SL interacts with a fetal self-ligand and that this causes the deletion of M54/SL expressing cells. This could explain the lack of fetal pre-B cells in both M54 SCID and M54 SCID/+ mice.
Although the transition of pro-B cells to the pre-B stage was not promoted in M54 SCID fetal liver, it was promoted in 3H9 SCID fetal liver. This differential effect of M54 and 3H9 did not correlate with an obvious difference in association of the M54 and 3H9 μ chains with SL chain, because μ-SL chain complexes were readily detectable in the pro-B cell fraction of both M54 and 3H9 SCID fetal liver. Moreover, a strong association of the M54 or 3H9 μ chain with SL chain would be expected as both tgs promoted pre-B cell development in SCID adult bone marrow. This indicates a strong association of M54 and 3H9 μ chains with SL chain because progression of bone marrow pro-B cells to the pre-B stage depends on expression of μ chains that efficiently associate with SL chain (11, 12). μ-SL chain association also would appear essential for the transition of most fetal pro-B cells to the pre-B stage because mice lacking the λ5 component of the SL chain not only show severely impaired development of pre-B cells in adult bone marrow (46) but also in fetal liver (47). As the differential effect of M54 and 3H9 on SCID fetal pre-B cell development cannot be attributed to a gross difference in efficiency with which M54 and 3H9 μ chains associate with SL chain, we suggest that it reflects a difference in pre-BCR specificity (M54/SL versus 3H9/SL).
μ/κ-tg SCID Mice.
In M54/Vκ8 SCID embryos, there was no evidence of an arrest at the pro-B cell stage. In the fetal liver of these embryos, B cells were detectable as early as day 17, 2 days earlier than in non-tg SCID/+ controls. Moreover, the percentage of B cells in M54/Vκ8 SCID fetal liver was normal, even though pre-B cells were virtually absent. It has been shown previously that Vκ8 is expressed early, at the pro-B cell stage (34), and that κ chains can substitute for SL chain in B cell development (48, 49). Therefore, we suggest that pairing of Vκ8 and M54 μ chains results in a receptor that is able to promote fetal B cell development because its specificity is sufficiently different from that of M54/SL. We conclude that the specificity of a pre-BCR (or prematurely expressed BCR) can influence the developmental fate of pro-B cells.
In sum, we suggest that expression of a pre-BCR may serve not only to signal the occurrence of a productive H chain gene rearrangement; it also may serve as the first checkpoint for elimination of cells with inappropriate H chains. For example, pro-B cells expressing a pre-BCR (or BCR) with auto-reactivity for a ligand in fetal liver or adult bone marrow could be deleted at the pro-B to pre-B transitional stage. Evidence for BCR-mediated cell deletion at the pro-B to pre-B transitional stage was reported earlier in a line of μκ-tg (Y-Sp6) SCID mice (34). A second checkpoint for deletion of auto-immune cells would be when cells normally express a BCR, at the pre-B to B transitional stage or immature B cell stage (41–45).
Acknowledgments
We thank Mark Schlissel for suggesting the cell transfer experiments and David Allman, Randy Hardy, Dietmar Kappes, Pamela Nakajima, Norman Ruetsch, David Wiest, and Martin Weigert for review of the manuscript. We especially thank N. Ruetsch for fluorochrome conjugation of antibodies and R. Hardy for providing a highly sensitive allophycocyanin-conjugated anti-B220 preparation. We also thank Roseanne Diehl for assistance in typing the manuscript. This work was supported by grants from the National Institutes of Health (CA06927 and CA04946) and an appropriation from the Commonwealth of Pennsylvania.
Abbreviations
- SCID
severe combined immune deficiency
- pre-BCR
pre-B cell receptor
- SL
surrogate light
- tg
transgene
- RAG
recombination activation gene
- RT-PCR
reverse transcriptase–PCR
- β2M
β2-microglobulin
References
- 1.Reth M. Annu Rev Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- 2.Cambier J C, Pleiman C M, Clark M R. Annu Rev Immunol. 1994;12:457–486. doi: 10.1146/annurev.iy.12.040194.002325. [DOI] [PubMed] [Google Scholar]
- 3.Melchers F, Haasner D, Grawunder U, Kalberer C, Karasuyama H, Winkler T, Rolink A R. Annu Rev Immunol. 1994;12:209–225. doi: 10.1146/annurev.iy.12.040194.001233. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 5.Reichman-Fried M, Hardy R R, Bosma M J. Proc Natl Acad Sci USA. 1990;87:2730–2734. doi: 10.1073/pnas.87.7.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitamura D, Roes J, Kuhn R, Rajewsky K. Nature (London) 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 7.Gong S, Nussenzweig M C. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 8.Shinkai Y, Rathbun G, Lam K-P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall S M, Alt F W. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 9.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura D, Rajewsky K. Nature (London) 1992;356:154–156. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- 11.ten Boekel E, Melchers F, Rolink A G. Immunity. 1997;7:357–368. doi: 10.1016/s1074-7613(00)80357-x. [DOI] [PubMed] [Google Scholar]
- 12.Kline G H, Hartwell L, Beck-Engeser G B, Keyna U, Zaharevitz S, Klinman N R, Jack H-M. J Immunol. 1998;161:1608–1618. [PubMed] [Google Scholar]
- 13.Bosma M J, Carroll A. Annu Rev Immunol. 1991;9:323–350. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- 14.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehlich A, Schaal S, Gu H, Kitamura D, Muller W, Rajewsky K. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- 16.Li Y-S, Hayakawa K, Hardy R R. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuler W, Weiler I J, Schuler A, Phillips R A, Rosenberg N, Mak T W, Kearney J F, Perry R P, Bosma M J. Cell. 1986;46:963–972. doi: 10.1016/0092-8674(86)90695-1. [DOI] [PubMed] [Google Scholar]
- 18.Erikson J, Radic M Z, Camper S A, Hardy R R, Carmack C, Weigert M. Nature (London) 1991;349:331–334. doi: 10.1038/349331a0. [DOI] [PubMed] [Google Scholar]
- 19.Grosschedl R, Weaver D, Baltimore D, Costantini F. Cell. 1984;38:647–658. doi: 10.1016/0092-8674(84)90259-9. [DOI] [PubMed] [Google Scholar]
- 20.Chang Y, Bosma G C, Bosma M J. Immunity. 1995;2:607–616. doi: 10.1016/1074-7613(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 21.Carmack C E, Camper S A, Mackle J J, Gerhard W U, Weigert M G. J Immunol. 1991;147:2024–2033. [PubMed] [Google Scholar]
- 22.Wasserman R, Li Y-S, Shinton S A, Carmack C E, Manser T, Wiest D L, Hayakawa K, Hardy R R. J Exp Med. 1998;187:259–264. doi: 10.1084/jem.187.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spanopoulou E, Roman C A J, Corcoran L, Schlissel L, Silver D P, Nemazee D, Nussenzweig M, Shinton S A, Hardy R R, Baltimore D. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 24.Yancopoulos G D, Desiderio S V, Paskind M, Kearney J F, Baltimore D, Alt F W. Nature (London) 1984;311:727–733. doi: 10.1038/311727a0. [DOI] [PubMed] [Google Scholar]
- 25.Perlmutter R M, Kearney J F, Chang S P, Hood L E. Science. 1985;227:1597–1601. doi: 10.1126/science.3975629. [DOI] [PubMed] [Google Scholar]
- 26.Wu G, Paige C. EMBO J. 1986;5:3475–3481. doi: 10.1002/j.1460-2075.1986.tb04672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong H D, Teale J M. J Exp Med. 1988;168:589–603. doi: 10.1084/jem.168.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yancopoulos G D, Malynn B A, Alt F W. J Exp Med. 1988;168:417–435. doi: 10.1084/jem.168.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan K M, Brodeur P H. EMBO J. 1989;8:2313–2320. doi: 10.1002/j.1460-2075.1989.tb08358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y-S, Wasserman R, Hayakawa K, Hardy R R. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 31.Cumano A, Paige C J, Iscove N N, Brady G. Nature (London) 1992;356:612–615. doi: 10.1038/356612a0. [DOI] [PubMed] [Google Scholar]
- 32.Winkler T H, Rolink A, Melchers F, Karasuyama H. Eur J Immunol. 1995;25:446–450. doi: 10.1002/eji.1830250221. [DOI] [PubMed] [Google Scholar]
- 33.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 34.Chang Y, Bosma M J. Int Immunol. 1996;9:373–380. doi: 10.1093/intimm/9.3.373. [DOI] [PubMed] [Google Scholar]
- 35.Marcu K B, Banerji J, Penncavage N A, Lang R, Arnheim N. Cell. 1980;22:187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- 36.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 37.Raff M C, Megson M, Owen J T, Cooper M D. Nature (London) 1975;259:224–226. doi: 10.1038/259224a0. [DOI] [PubMed] [Google Scholar]
- 38.Chang Y, Bosma M J, Bosma G C. J Exp Med. 1999;189:1295–1305. doi: 10.1084/jem.189.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corcos D, Dunda O, Butor C, Cesbron J-Y, Lores P, Bucchini D, Jami J. Curr Biol. 1995;5:1140–1148. doi: 10.1016/s0960-9822(95)00230-2. [DOI] [PubMed] [Google Scholar]
- 40.Shaffer A L, Schlissel M S. J Immunol. 1997;159:1265–1275. [PubMed] [Google Scholar]
- 41.Nemazee D A, Burki K. Proc Natl Acad Sci USA. 1989;86:8039–8043. doi: 10.1073/pnas.86.20.8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartley S B, Cooke M P, Fulcher D A, Harris A W, Cory S, Basten A, Goodnow C C. Cell. 1993;72:325–335. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 43.Tiegs S L, Russell D M, Nemazee D. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gay D, Saunders T, Camper S, Weigert M. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, Nagy Z, Radic M Z, Hardy R R, Huzar D, Camper S A, Weigert M. Nature (London) 1995;373:252–255. doi: 10.1038/373252a0. [DOI] [PubMed] [Google Scholar]
- 46.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 47.Ceredig R, ten Boikel E, Rolink A, Melchers F, Andersson J. Int Immunol. 1998;10:49–59. doi: 10.1093/intimm/10.1.49. [DOI] [PubMed] [Google Scholar]
- 48.Pelanda R, Schwers S, Sonoda E, Torres R M, Nemazee D, Rajewsky K. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- 49.Papavasiliou F, Jankovic M, Nussenzweig M C. J Exp Med. 1996;184:2025–2030. doi: 10.1084/jem.184.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]