Abstract
Objective
This paper reports on the alignment between two large ontologies of anatomy: the Foundational Model of Anatomy (FMA) and the representation of anatomical structures in SNOMED CT. The objective of this study is to investigate the compatibility between a reference ontology of anatomy (the FMA, 75,019 concepts) and a representation of anatomy created for use in clinical applications (SNOMED CT, 30,933 anatomical concepts).
Methods
The alignment first identifies shared concepts lexically. The presence of shared relations across ontologies is then used to validate the mappings structurally.
Results
8,228 mappings were identified by lexical methods, of which over 97% were supported by structural evidence. No evidence was found for 0.5% of the mappings and 2.5% received negative evidence.
Conclusions
Despite important differences in coverage and knowledge representation between the FMA and SNOMED CT, we have not noticed any major discrepancies in their representation of anatomical entities.
INTRODUCTION
Many representations of anatomy have been developed for various purposes. While some of them are mere lists of names for anatomical entities (e.g., Terminologica Anatomica), others are full-fledged ontologies, organizing anatomical entities in hierarchies (isa, part of) in order to support reasoning (e.g., Foundational Model of Anatomy, SNOMED CT). Because they include information about relations among anatomical entities, anatomical ontologies can be aligned accurately. In previous work, we have developed methods for aligning such ontologies, based not only on the lexical resemblance of concept names across ontologies, but also on the similarity of relations among these concepts across ontologies [1]. We have applied this method to several pairs of anatomical ontologies and validated it against a gold standard constituted manually [2].
While the general framework of this study is that of ontology alignment, our interest here goes beyond the alignment itself. No specific alignment method has been created for this study. Rather, we have reused the techniques developed for aligning other anatomical ontologies. (For a survey of alignment techniques, the interested reader is referred to [3].) The contribution of this paper is to exploit the alignment of anatomical entities in two ontologies for analyzing the differences between these ontologies. Additionally, this paper is an attempt to reflect on the consequences of these differences on the compatibility between these ontologies, as well as on the alignment itself.
The objective of this study is to apply ontology alignment techniques to two ontologies of anatomy developed for different purposes and analyze some of the differences in their representation of anatomical entities. The two ontologies under investigation are the Foundational Model of Anatomy, created as a reference, purpose-independent ontology of anatomy and SNOMED CT, a large clinical vocabulary of which anatomy is one component, along with clinical findings, medical procedures, pharmaceutical products and many other aspects of clinical medicine.
MATERIALS
The Foundational Model of Anatomy (FMA)1 is an evolving ontology that has been under development at the University of Washington since 1994 [4]. Its objective is to conceptualize the physical objects and spaces that constitute the human body. The underlying data model of the FMA is a frame-based structure implemented with Protégé. 75,019 concepts cover the entire range of macroscopic, microscopic and subcellular canonical anatomy. In addition to preferred terms (one for each concept), 53,451 synonyms are provided (up to 13 per concept). For example, there is a concept named Uterine tube and its synonym is Oviduct. Because single inheritance is one of the modeling principles used in the FMA, every concept (except for the root) stands in a unique is-a relation to other concepts. Additionally, concepts are connected by five kinds of part of relationships (e.g., part of, constitutional part of, regional part of). For alignment purposes in this study, we consider as only one part of relationship (with has part as its inverse) the various kinds of partitive relationships present in the FMA. The version used in this study is v1.3.0 dated of October 13, 2005.
SNOMED Clinical Terms® (SNOMED CT®)2, is an evolving clinical health care terminology developed by the College of American Pathologists. The goal of SNOMED CT is to provide “a common language that enables a consistent way of capturing, sharing and aggregating health data across specialties and sites of care”. Chief among its applications are electronic medical records. While description logics-based technologies are used for its development, SNOMED CT is distributed in relational format through the Unified Medical language System® (UMLS®)3. The version used in this study (July 2005, from UMLS 2005AC) comprises some 300,000 concepts, of which 30,933 pertain to anatomical structures. Concept names – descriptions in SNOMED CT parlance – include one fully specified term (e.g., Entire skin of flank (body structure)) and synonyms (up to 37 per concept, e.g., Skin of side of abdomen). Two kinds of relationships link anatomical concepts in SNOMED CT: isa and part_of. More precisely, SNOMED CT uses a representation of anatomical entities based on Structure-Entire-Part (SEP) distinctions [5, 6]. For example, the right hand (Entire right hand) is represented as follows:
Entire right hand isa Entire hand
Entire right hand isa Structure of right hand
Entire right hand part_of Entire right upper extremity
Although not entirely intuitive, this representation offers interesting computational properties derived from the reification of part of relations. Namely, traversing the isa link yields both the concepts subsumed by a given anatomical entity and the concepts corresponding to parts of this anatomical entity. We used this features for extracting the set of all anatomical concepts in SNOMED CT as the isa descendants of the high-level concept Biological structure.
For reasons explained in detail in the discussion section, the counterpart in SNOMED CT of the anatomical entities in the FMA is represented by the Entire concepts in the SEP triples.
METHODS
The method used for aligning the Foundational Model of Anatomy (FMA) and SNOMED CT was originally developed by the authors for aligning the FMA and GALEN [1] and can be summarized as follows. Concept names and relations are extracted from each ontology. In the lexical approach, additional synonyms are collected. All names are normalized and compared across ontologies. Lexically similar names form the basis for identifying equivalent concepts. Structural similarity (e.g., shared relations to other equivalent concepts) is required for concepts to be aligned.
Lexical alignment
The lexical alignment identifies shared concepts across systems lexically through exact match and after normalization. For example, the terms neutrophil in FMA and polymorphonuclear leukocyte in SNOMED CT match exactly because polymorphonuclear leukocyte and neutrophil are synonyms in the FMA. Other examples of matches include the FMA term Intervertebral disk, T10-T11 and the SNOMED CT term Intervertebral disc, T10-T1. Here, normalization eliminates minor differences in terms, such as spelling variants (disk/disc).
While the simpler term Right hand is a synonym for both Entire right hand and Structure of right hand, simple terms are not systematically provided for both Entire and Structure concepts. In order to maximize the chances of identifying a lexical match in SNOMED CT and to ensure consistent mapping to the Entire concepts, we systematically created the simpler synonyms from Entire and Structure concepts, as necessary. For example, we added the term Right kidney – originally a synonym for Right kidney structure only – as a synonym for Entire right kidney also.
Concepts exhibiting similarity at the lexical level across systems are called anchors, as they are going to be used as reference concepts in the structural alignment.
Validation by structural similarity
In the structural validation of the lexical alignment, the first step is to acquire the semantic relations explicitly represented in each system. In order to facilitate the comparison of relations across systems, the transitive closure of isa relations is computed in each system, as well as that of part of relations. With these semantic relations, the structural alignment identifies structural similarity among anchors across systems. Structural similarity, used as positive structural evidence, is defined by the presence of at least one common hierarchical relation among anchors across systems, e.g., <c1, part of, c2> in one system and <c1’, part of, c2’> in another where {c1, c1’} and {c2, c2’} are anchors across systems. For example, the anchor concepts neutrophil in the FMA and polymorphonuclear leukocyte in SNOMED CT, presented earlier, received positive structural evidence because they share hierarchical links to other anchors across systems. Neutrophil is related to granular leukocyte (isa) and to hematopoietic system (part of). These relations from the FMA mirror relations among equivalent concepts in SNOMED CT. One minor difference is that the relation of neutrophil to hematopoietic system is direct in SNOMED CT and indirect [through blood (part of)] in the FMA. The structural validation is performed automatically.
While looking for structural similarity, structural discrepancies can also be detected, resulting in negative evidence for a given lexical match. For example, although joint(s) is a synonym for both Set of joints in the FMA (joints) and Entire joint in SNOMED CT (joint), these two concepts do not constitute a mapping because they share different, incompatible relations to articular system (Articular system isa Set of joints in the FMA and Entire joint part of Entire articular system structure in SNOMED CT).
RESULTS
Lexical alignment
3,979 synonyms were generated in SNOMED CT, i.e., 2,744 for Entire concepts and 1,235 for Structure concepts. 8,228 lexical matches were identified, accounting for about 11% of all FMA concepts and 27% of all SNOMED CT concepts.
Structural validation
The vast majority (over 97%) of the 8,228 lexical matches is supported by structural evidence. Only 41 of them (0.5%) are rejected for lack of structural evidence and 204 matches (2.5%) are rejected because of conflicting relations to other anchors.
DISCUSSION
Aligning anatomical entities in the FMA and SNOMED CT enables us to analyze some of the differences between the two ontologies in terms of their consequences on the alignment (knowledge representation, terminology, coverage).
Differences in knowledge representation and terminological differences
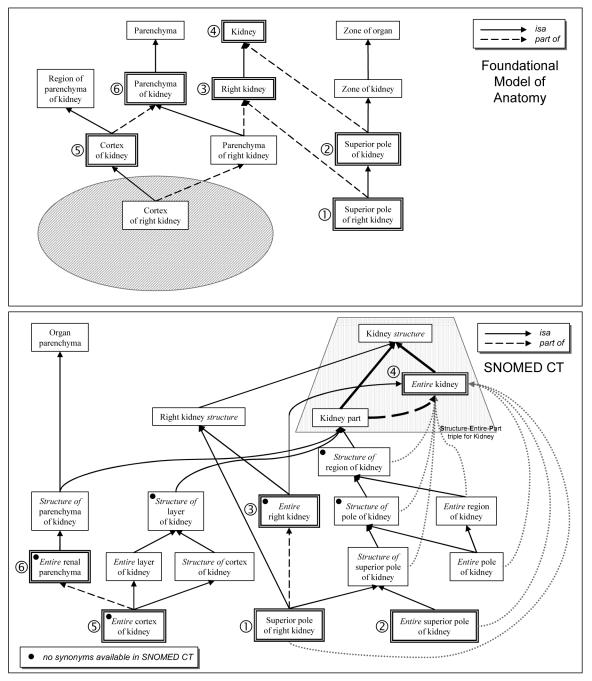
SNOMED CT’s representation of anatomy relies on the so-called Structure-Entire-Part (SEP) triples. The SEP representation was created by Schultz & al. [6] to support mereological reasoning in medical ontologies [7]. Three concepts are used to represent each anatomical entity. The Entire concept represents the entire anatomical entity. The Part concept results from the reification of the partitive relation and represents any parts of the entity. Finally, the Structure concept subsumes the other two and represents the entity or any of its parts. In addition to subsumption, there exists a mereological relation (part of) between the Part and the Entire concepts. The SEP triple for Kidney is shown in Figure 1. In fact, there are not always three concepts for each entity in SNOMED CT, but most often two: the Entire and the Structure. For example, there is no such concept as Right kidney part although there exist Entire right kidney and Right kidney structure.
Figure 1.
Representation of Kidney in the FMA and SNOMED CT. (The concepts identified by numbers correspond to mappings across ontologies)
Many Structure and Entire concepts share synonyms. For example, the name Kidney is common to both Kidney structure and Entire kidney. As a consequence, a large number of FMA names are ambiguous in SNOMED CT, resulting in multiple lexical matches. We resolve the ambiguity by associating the FMA term X to the SNOMED CT term Entire X rather than Structure of X (or X structure). This simple rule allowed for the disambiguation of 5,196 multiple matches. Of note, the sharing of names between Structure and Entire concepts is by far not systematic. For example, in Figure 1, no synonyms are provided in SNOMED CT for the six concepts denoted by a black dot. Entire right kidney had no synonyms, while Right kidney is a synonym for Right kidney structure. As a consequence, the term Right kidney in the FMA could not have been mapped to the term Entire right kidney in SNOMED CT, had we not created its simpler synonym Right kidney. Moreover, the term Right kidney in the FMA would have been mapped inaccurately to Right kidney structure instead.
Although using description logics for its development, SNOMED CT is distributed in relational format, with all mereological inferences precomputed. For example, in the representation of kidney shown in Figure 1, seven of the eight part of relations to Entire kidney are actually inherited from Kidney structure part of Entire kidney. Added to the presence of “redundant” concepts, inherited partitive relations make the representation in SNOMED CT look somewhat cluttered compared to the FMA. However, these relations are not detrimental to the alignment process. In fact, in order to maximize the chances of finding structural evidence to support lexical matches, we also compute the transitive closure of part of relations. In the case of SNOMED CT, these relations already exist in the ontology.
Interestingly, the reification of partitive relations realized by the Part concepts in the SEP representation is not specific to SNOMED CT. 574 concepts in the FMA have names of the form “Subdivision of X” (e.g., Subdivision of pharynx). For each isa descendant Y of such concepts, the alignment process creates an explicit relation Y part of X whenever such a relation does not already exist.
Differences in coverage
The number of anatomical concepts in the two ontologies (75,019 in the FMA vs. 30,933 in SNOMED CT) suggests that their coverage must differ significantly. On the one hand, the difference seems even larger if we take into account the concepts created in SNOMED CT purely for knowledge representation purposes, because there is no correspondence in the FMA for most of these concepts. In order to get a rough estimate of the concept “redundancy” due to the SEP representation, we counted in SNOMED CT the unique number of anatomical concepts whose names contain structure, entire, and part as a proper substring: 9,099, 8,459 and 647, respectively, for a total of 17,964 unique concepts (some names may contain several of these words). Assuming Entire and Part concepts are “redundant” with some Structure concepts, these 17,964 unique concepts correspond at most to 9,099 distinct anatomical structures.
On the other hand, because of its precoordinated nature, the FMA creates concepts for all structures. Conversely, SNOMED CT concepts can be created by coordinating existing concepts. As illustrated in Figure 1, Cortex of right kidney is represented only in the FMA. In SNOMED CT, an equivalent concept would result from refining the laterality of Cortex of kidney with Right, one of the allowable values for laterality. In summary, the difference in number of concepts between the FMA and SNOMED CT does not reflect adequately differences in coverage.
As a reference, purpose-independent ontology, the FMA essentially restricts its representation of anatomy to the structural perspective and to canonical anatomy. In contrast, SNOMED CT represents both normal and pathological structures (e.g., tumors such as glioblastoma), as well as non-pathological, yet non-canonical structures (e.g., Gravid uterus structure, Placental villus, and Sixth branchial cleft). Additionally, SNOMED CT inherited from its predecessor the tradition of accommodating veterinary medicine and represents non-human anatomical structures, including Paw, Eighteenth rib and Pectoral fin. Most importantly, the representation of anatomy in SNOMED CT is oriented toward its use in clinical medicine. Topography, for example, includes acupuncture points (e.g., Huatuochiachi C1), electrocardiograph lead sites (e.g., Lead site V1) and other clinical references (e.g., Diaper area, Diabetic Retinopathy Study field 1) in addition to the reference anatomical landmarks. Purposely absent from the FMA, but represented in SNOMED CT is the functional perspective on anatomy, with concepts representing the type of movement in which the muscle (or group there of) participates, such as Extensor muscle of hand and Flexor of shoulder joint.
Compatibility
Despite the important differences in their representation mechanisms and coverage highlighted above, we have not noticed any major discrepancies in the representation of anatomical entities between the FMA and SNOMED CT. A finer analysis involving anatomists, ontologists and knowledge representation specialists would be required to confirm this finding. Meanwhile, it seems that a mapping to the FMA was identified for a large part of the anatomical entities corresponding to human canonical anatomy in SNOMED CT. The coverage provided by the FMA remains finer-grained. However, a large proportion of concepts in the FMA (over 40%) differ from other concepts only by laterality (e.g., Left ligament of wrist vs. Ligament of wrist). Rather than representing with precoordinated terms those fine-grained concepts exhibiting laterality distinctions, SNOMED CT makes it possible for users to create them on the fly. However, SNOMED CT would certainly benefit from a more consistent representation of the concept names, especially between Entire and Structure concepts.
It is beyond the scope of this paper to fully evaluate the mapping between the FMA and SNOMED CT. However, based on previous evaluations of our alignment technique, we are reasonably confident in the quality of the mappings we identified. This study also showed that refining our alignment techniques for these two ontologies (e.g., creating all synonyms for Entire and Structure concepts) was critical to the identification of additional mappings and to the accuracy of the alignment.
Acknowledgements
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM) and by the Natural Science Foundation of China (No.60496324), the National Key Research and Development Program of China (Grant No. 2002CB312004), the Knowledge Innovation Program of the Chinese Academy of Sciences, and MADIS of the Chinese Academy of Sciences, and Key Laboratory of Multimedia and Intelligent Software at Beijing University of Technology. Thanks for their support to Cornelius Rosse, José Mejino and Todd Detwiler for the Foundational Model of Anatomy.
Footnotes
References
- 1.Zhang S, Bodenreider O. Aligning representations of anatomy using lexical and structural methods. AMIA Annu Symp Proc. 2003:753–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Bodenreider O, Hayamizu TF, Ringwald M, de Coronado S, Zhang S. Of mice and men: Aligning mouse and human anatomies. Proc AMIA Symp. 2005:61–65. [PMC free article] [PubMed] [Google Scholar]
- 3.Staab S, Studer R, editors. Tools for mapping and merging ontologies. Handbook on Ontologies: Springer-Verlag; 2004. pp. 365–384. [Google Scholar]
- 4.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Schulz EB, Price C, Brown PJ. Symbolic anatomic knowledge representation in the Read Codes version 3: structure and application. J Am Med Inform Assoc. 1997;4(1):38–48. doi: 10.1136/jamia.1997.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz S, Romacker M, Hahn U. Part-whole reasoning in medical ontologies revisited--introducing SEP triplets into classification-based description logics. Proc AMIA Symp. 1998:830–4. [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz S, Hahn U. Part-whole representation and reasoning in formal biomedical ontologies. Artif Intell Med. 2005;34(3):179–200. doi: 10.1016/j.artmed.2004.11.005. [DOI] [PubMed] [Google Scholar]