Abstract
Electronic health records are designed to provide online transactional data recording and reporting services that support the health care process. The characteristics of clinical data as it originates during the process of clinical documentation, including issues of data availability and complex representation models, can make data mining applications challenging. Data preprocessing and transformation are required before one can apply data mining to clinical data. In this article, an approach to data preparation that utilizes information from the data, metadata and sources of medical knowledge is described. Heuristic rules and policies are defined for these three types of supporting information. Compared with an entirely manual process for data preparation, this approach can potentially reduce manual work by achieving a degree of automation in the rule creation and execution. A pilot experiment demonstrates that data sets created through this approach lead to better model learning results than a fully manual process.
Introduction
Modern electronic health records (EHR’s) are designed to capture and render clinical data during the health care process. Using them, health care providers can enter and access clinical data when it is needed. Through the presence of digital data, EHR’s can incorporate decision support technologies to assist clinicians in providing better care. When adequate data is recorded in an EHR, data mining technologies can be used to automatically extract useful models and can assist in constructing the logic for decision support systems.
However, because the main function of EHR’s is to store and report clinical data collected for the purpose of health care delivery, the characteristics of this data may not be optimal for data mining and other data analysis operations[1]. One challenge in applying data mining to clinical data is to convert data into an appropriate form for this activity. Data mining algorithms can then be applied using the prepared data. The adequacy of data preparation often determines whether this data mining is successful or not.
In this article, we propose a data preparation framework for transforming raw transactional clinical data to well-formed data sets so that data mining can be applied. In this framework, rules are created according to the statistical characteristics of the data, the metadata that characterizes the host information systems and medical knowledge. These rules can be used for data preprocessing, attribute selection and data transformation in order to generate appropriately prepared data sets. In contrast with an entirely manual process for data preparation, this approach can potentially reduce human work by achieving a certain degree of automation in the rule creation and execution.
Development Motivation
As a part of the development effort for a new decision support system, we are creating tools that can help identify medical problems for physicians who would like to maintain an electronic problem list. One approach is to use data mining technologies to develop an expert system that can inspect raw clinical data and propose problems to clinicians as they maintain this problem list. The goal of this expert system is to assist physicians in identifying all medical problems and to facilitate the completeness and timeliness of the medical problem list. We have previously tested the user interface for “proposed” problems in an application where the target problems were extracted from medical documents using natural language processing techniques[2]. This work will extend that model to allow the prediction of problems based on the clinical data available in the EHR.
For the development of this expert system, we are using a clinical data repository where data is stored in a raw format similar to that used in the online transactional system where the data was captured. Determining an approach to processing this data into an appropriate form for data mining has been an important challenge. Thus, we have developed a systematic way of preparing data in an effort to save manual work and to get better results from the data mining process.
Issues of Raw Transactional Clinical Data
Before one can use any automatic model learning method to extract useful models from data, he/she must process the data into a form that is acceptable to the learning method. The “flattened table” format is most common and is required for most methods. In the flattened table format, each row represents an instance for training and/or testing a model (often containing relevant data for an individual patient); each column represents the values for a variable across the instances. Despite the fact that this model is simple and commonly used in data analysis, it is not the format stored in typical EHR’s. Due to the characteristics of clinical practice and of the data structures required in an EHR, some form of data transformation is invariably necessary to convert data from its original format to a flattened table. The challenges include:
Storage structure
The database structure of raw clinical data is usually designed to support an online transactional system, which is optimized for patient-based transactions, e.g., with indexes and structures specialized to single-patient transactions. While these database structures work effectively with transactions involving the data of individual patients, they typically are not as effective with trans-population queries. For data mining and statistical analysis, population data must ultimately be rendered as a flattened table.
Poor Data quality and Inconsistent Representations
Data in a raw clinical data repository can be of poor quality. Outliers due to entry errors are commonly found. Inconsistent representation of data can exist, especially if more than one model for expressing a specific meaning exists (e.g., for abdominal rebound pain, one application might enter it as a specific nominal variable with value “YES” and another might provide only the option of entering “abdominal pain” as free text). In addition, the data type for data in databases does not always reflect the true data type. For example, a column of the numeric data type in databases can represent a nominal or ordinal variable encoded in numbers instead of a true continuous variable (as in deep tendon reflexes represented as the numerics 0, 1, 2, 3, and 4). When one evaluates variables based on parameters such as mean and variance, he/she must consider this type of data presentation.
Too many variables
For algorithms where the computational complexity is more than linear, the time required may become infeasible as the number of variables grows. However, the number of variables stored in EHR for each patient can be greater than 1000, which makes many algorithms impractical in terms of the time they would take.
Missing data elements
Clinical data elements often are not collected for all data required for analysis. Some data elements are not collected because of omission, irrelevance, excess risk, or inapplicability in a specific clinical context. For some model learning methods such as logistic regression, a complete set of data elements may be required. Even when methods that accept missing values are used, the fact that the data was not collected may have independent information value and should not be ignored (i.e., data is often not missing in a random manner[3]). Methods of data imputation[3] and of modeling the missing data are necessary to cope with this issue.
Data Warehousing Issues
Ideally, the data warehousing process should mitigate these issues and make it easier for researchers and data analysts to acquire the data and information they need. A data warehouse for clinical data should render the data in appropriate structures, provide metadata that adequately records syntax/semantics of data and reference pertinent medical knowledge. However, existing clinical data warehouses typically fail to support these functions perfectly. Some data may simply exist in a format similar to that found in the EHR.
The Manual Process of Data Preparation
When one wants to extract useful models from data for a specific problem (e.g., a predictive model to detect patients with pneumonia), he/she would usually begin by consulting medical knowledge sources for relevant clinical variables and then would explore the data source for data elements that represent the clinical variables. This manual process requires mapping from clinical terms to data elements in the EHR. Sometimes the clinical term is abstract and can only be represented by combinations of data elements. For example, the term “inflammation” is a condition inferred from a combination of vital sign abnormalities and some local and laboratory findings. “Inflammation” rarely exists as a single data element in the EHR. The mapping, including abstraction, involves knowledge of medicine and of the data organization in the EHR.
This manual process to define and prepare data sets, though intuitive, has two disadvantages. One is that it is labor-intensive and demands knowledge of both medicine and the data organization of the EHR. The ambiguity between clinical terms and data elements is sometimes difficult to resolve. The other disadvantage is that the related variable set is limited by available domain knowledge. If a potentially useful prediction model uses a variable that is not known to be related to the target problem, this model cannot be found using the manual process.
Automation Using Helpful Information
In order to reduce the challenges discussed above, we have design a data preparation framework that utilizes information from three areas – raw data, available metadata and domain knowledge. This framework is used by applying a group of heuristic rules and policies. We believe that this framework can be particularly effective in reducing the part of the manual work that demands domain knowledge for data preparation. The goal remains to provide relevant data sets that will lead to good model learning results.
Material and Methods
This study was conducted using data extracted from the enterprise data warehouse (EDW) of Intermountain Health Care (IHC) in Salt Lake City. The data was captured during routine clinical care documented in the HELP[4] hospital information system. IHC has established a working process that duplicates data from the HELP system to a data mart in the EDW called the “HELP” data repository.
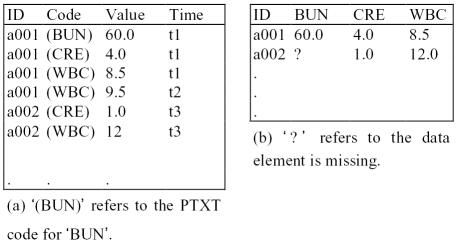
Although the data has been transformed from its original format into relational database tables, the organization of data is kept by using the “variable-value pair” data presentation model (Figure 1). The characteristics of this data source are similar to the original online transactional data in content and structure.
Figure 1.
(a) “Variable-value pair” data presentation model. The example is simplified by excluding supporting data such as “specimen type”. The definition in parentheses represents the actual, 8-byte, PTXT code stored in the database (b) flattened table
Clinical data in the HELP system is encoded using a data dictionary called “PTXT’. Although textual descriptions are available for most PTXT codes, they do not explicitly describe the real variable type and its possible values. For example, a numeric data type may represent an enumerated categorical variable instead of a true numeric variable. Sometimes a code from the dictionary represents a value instead of a variable; the real variable is hidden in the code hierarchy.
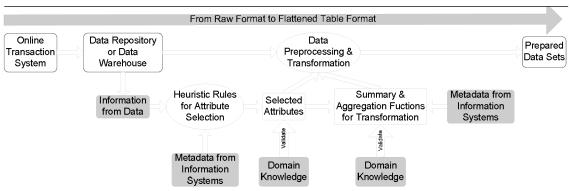
Although the characteristics of this data source provide challenges in converting data into the flatten table format, supplemental information sources can be used to support this effort. In our data preparation framework, relevant information is extracted from three sources – data, metadata and domain knowledge (Table 1). Rules and policies were designed in each area to support data preparation, including prescreening data elements, transforming data and providing summary and aggregation functions (Figure 2).
Table 1.
Examples of helpful information for preparing clinical data for data mining
| Source Type | Examples |
|---|---|
| Data | Descriptive statistics derived from the data set, including mean, variance, the number of distinctive values, the number of occurrences, etc. |
| Comparative statistics between case and control groups, such as χ2 test, t-statistics, information gain methods | |
| Metadata | Code descriptions provided by the information system |
| Medical Knowledge | The literature |
| Experts | |
| Medical knowledge base |
Figure 2.
Data preparation framework
Pilot Experiment
To compare data preparation using this framework to the entirely manual process, we conducted a pilot experiment. We developed a system to detecting patients who were admitted to the hospital with pneumonia. The data set included data of patients who were discharged from the hospital with pneumonia as primary diagnosis as well as a group of control patients. Both groups were sampled from patients admitted to LDS Hospital from the year 2000 to 2004.
The manual approach is to acquire variables relevant to pneumonia according to domain knowledge and the medical literature. Keyword searches were used on the code description field to find a list of candidate data codes. The candidate codes were inspected and the most suitable codes were chosen. The earliest observed value for each code was selected as the summary value for the chosen period. Each time no value was found for an instance of a variable, the variable was discretized and a state called ‘missing’ was added to it. By using this process, a data set in flattened-table format could be created from the original data.
In our experimental approach, two types of heuristic rules were used to select variables (detailed in Figure 3). One is to prescreen data elements based on their statistical characteristics and their gross categorization in the data dictionary. This allows us to select data subsets that are relevant to the clinical model that we are developing. The second is to select data elements able to differentiate the specific clinical problem (in this case pneumonia) according to comparative statistics (i.e., χ2 and two-sample t test) calculated from the test and control groups across all candidate variables. The candidate variable list was then manually inspected to remove obviously irrelevant variables. A second flattened-table data set was created.
Figure 3.
Steps of preprocessing data for pneumonia detection model
Comparison of Data Preparation Processes
The data sets acquired by the data preparation framework and the manual process were sent to two Bayesian[5] network learning modules – one is naïve Bayesian learner and the other is the WinMine[6] structure and parameter learner. The performance of the four classifiers derived from these two data sets was compared using the area under receiver operating characteristic (ROC) curves[7] combined with bootstrapping[8] procedures using 500 iterations. The 95% confidence intervals for the difference of the areas under ROC curves (AROC) between these two data preparation approaches were extracted from the 500 iterations.
Pilot Results
The numbers of patients in the case and control groups were 1521 and 1376 respectively. The variables that were extracted for the pneumonia prediction model by these two processes are listed in Table 2. The comparison of the pilot results produced by these two data preparation processes are shown in Table 3. The two 95% confidence intervals of the difference of AROC are above zero, indicating that the difference is statistically significant (α=0.05). The result show the two tested model learning algorithms performed better with the data set prepared by the framework.
Table 2.
Variables extracted by two approaches.
| Manual process | Data preparation framework |
|---|---|
| PO2 | CO2 (Serum) |
| PCO2 | Band% |
| Band% | Seg% |
| Eos% | BobyTemp(Ear Probe) |
| Neutrophil% | Resp.Rate |
| Seg% | HeartRate |
| WBC | Age |
| SpO2 | WBC |
| BodyTemp | ChestXRay Order(*p) |
| HeartRate | SpO2(*p) |
| Resp.Rate | Cough(*p) |
| Ronchi | Suspected Breathing Sound(*p) |
| Rales | Wheeze(*p) |
| Crackles | CO2serum(*p) |
| Productive cough | WBC(*p) |
| WBC differential count(*p) |
p: dichotomous variable of presence or absence.
Table 3.
Comparison of the areas under ROC of different approaches to provide data sets
| Naïve Bayes | WinMine | |
|---|---|---|
| Manual process | 0.9792 | 0.9833 |
| Data preparation framework | 0.9846 | 0.9905 |
| 95%C.I. of difference in AROC | (0.0038~0.0067) | (0.0050~0.0094) |
Discussions
Generalization
The process of data mining often depends on the domain where it is applied, yet general principles remain. Before one can use any model learning algorithm, he/she must preprocess and transform data into an appropriate form. The preparation process varies depending on the characteristics of the original data and the goals of data mining. Thus, different clinical data sources and different clinical problems may require different approaches to data preparation.
Nevertheless, the approach described here should be applicable to many clinical data sources. Rules based on common data characteristics (such as the mean of numeric values and the number of distinctive values in data) will work in the majority of cases. However, rules and policies that are defined based upon features specific to an EHR may not be directly applicable to other EHR’s.
Potential Advantages of the Proposed Framework
The execution of the rules based on data characteristics can reduce the number of candidate data elements so manual work can be reduced. Also, the resulted data set will be more consistent than through an entirely manual process.
In addition, the proposed framework uses data characteristics to select variables. This avoids the mapping process from clinical terms to data elements although manual validation may still be required.
Although classical feature selection algorithms are often designed to deal with similar variable selection problems, they usually require a well-prepared data set, which is rarely available in raw clinical data. Our approach applied fundamental descriptive statistics early in the process and can be more flexible in dealing with not so well-formed raw clinical data.
Challenges of Implementation
The objective of this framework is to reduce manual work. The extra manual work would be saved once the screening rules have been established. However, the definition of rules and policies requires knowledge of medicine and of data characteristics in the information system. Adequate metadata provided by information systems can help in this process. Domain knowledge is also required in result validation. Defining the rule and validating the data set are usually multi-iteration rather than single-passed. Nevertheless, many rules are reusable within the same EHR system. Thus, the proportion of work will decrease as the scale of the project grows.
Conclusions
In this article, we proposed a data preparation framework for converting raw clinical data to a format that is acceptable to model learning algorithms. In this framework, information is categorized into three main areas and rules and policies can be made according to the information. By using these rules and policies, the manual work required for this process can be reduced and information from various sources can be used in a systematic way. The pilot experiment result also suggested that better model learning could be achieved by using this framework.
References
- 1.Cios KJ, Moore GW. Uniqueness of medical data mining. Artif Intell Med. 2002;26:1–24. doi: 10.1016/s0933-3657(02)00049-0. [DOI] [PubMed] [Google Scholar]
- 2.Meystre S, Haug PJ. Automation of a problem list using natural language processing. BMC Medical Informatics and Decision Making. 2005;5:30. doi: 10.1186/1472-6947-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Little RJA, Rubin DB. Statistical analysis with missing data. 2. Wiley-Interscience; 2002. [Google Scholar]
- 4.Gardner RM, Pryor TA, Warner HR. The HELP hospital information system: update 1998. Int J Med Inf. 1999;54(3):169–82. doi: 10.1016/s1386-5056(99)00013-1. [DOI] [PubMed] [Google Scholar]
- 5.Jensen FV. Bayesian networks and decision graphs. New York: Springer-Verlag; 2001. [Google Scholar]
- 6.Chickering DM. The WinMine Toolkit. Redmond, WA: Microsoft; 2002. [Google Scholar]
- 7.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 8.Efron B, Tibshirani R. Improvements on cross-validation: the .632+ bootstrap method. Journal of American Statistical Association. 1997;92:548–60. [Google Scholar]