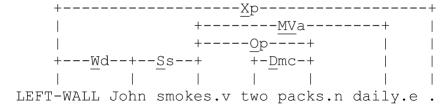Abstract
Semantic category recognition (SCR) contributes to document understanding. Most approaches to SCR fail to make use of syntax. We hypothesize that syntax, if represented appropriately, can improve SCR. We present a statistical semantic category (SC) recognizer trained with syntactic and lexical contextual clues, as well as ontological information from UMLS, to identify eight semantic categories in discharge summaries. Some of our categories, e.g., test results and findings, include complex entries that span multiple phrases. We achieve classification F-measures above 90% for most categories and show that syntactic context is important for SCR.
Introduction
Rapid search and retrieval of clinical records can be empowering. However, a large portion of clinical records are in free text; extracting information from these records requires linguistic processing. Here, we focus on semantic category recognition (SCR); SCR is a subset of information extraction (IE) and is a first step in the semantic interpretation of documents.
MedLEE is a system for IE in medical discharge summaries.1 This system uses a lexicon for the semantic types of words and phrases, parses text using a grammar, and maps phrases to standardized semantic frames for medical findings and diseases.
The UMLS Metathesaurus is frequently used for concept recognition. MetaMap2 identifies candidate phrases through shallow parsing and applies a greedy search algorithm to map these phrases (or their substrings) to Metathesaurus concepts. Delbecque3 takes a similar approach. Some systems map phrases first to UMLS semantic types and then to more specific semantic categories, e.g., diagnoses and procedures in Long4. In addition to dictionaries, some IE approaches rely on context. The Badger5 text analysis system uses a semantic lexicon and a dictionary of syntactic and contextual rules to recognize information concerning diagnoses and symptoms. Averbuch uses context to identify negative/positive instances of various symptoms.6
We present a statistical SC recognizer that resembles the aforementioned systems in various ways. Like Long4, we rely on the UMLS knowledge-base. However, we enrich information from UMLS with lexical and syntactic context. We aim to identify diseases, signs and symptoms (referred to as symptoms), treatments, diagnostic tests (referred to as tests), results, dosage information (referred to as dosages), abusive substances (referred to as substances), and medical practitioners (referred to as practitioners) in medical discharge summaries; we employ support vector machines (SVMs) as a means to this end. Our SC recognizer differs from previous work in several ways: while previous work focused on extracting semantic categories that frequently consist of single words or simple noun phrases, e.g., diseases, we also include complex categories, e.g., results, which consist of complex phrases and entire clauses. In addition to the surface features and lexical n-grams (uninterrupted strings of words) frequently employed for SCR, we use syntactic dependencies extracted from the Link Grammar Parser and show that this syntactic information contributes significantly to SCR even in ungrammatical medical discharge summaries.
Data and Methods
We experimented with a collection of 48 summaries (containing a total of 5,166 sentences) obtained from various medical departments. Based on the advice of two doctors, we defined eight semantic categories related to medical discharge summaries. We mapped these categories to UMLS semantic types as follows:
Diseases include the UMLS semantic types Pathologic Functions, Disease or Syndrome, Mental or Behavioral Dysfunction, Cell or Molecular Dysfunction, Congenital Abnormality, Acquired Abnormality, Injury or Poisoning, Anatomic Abnormality, Neoplastic Process, and Virus/Bacterium.
Treatments include the semantic types Therapeutic or Preventive Procedure, Medical Device, Steroid, Pharmacologic Substance, Biomedical or Dental Material, Antibiotic, Clinical Drug, and Drug Delivery Device.
Substances refer to abusive drugs or substances, e.g., narcotics and tobacco. The closest UMLS semantic type is Hazardous or Poisonous Substance.
Dosages include the amount and the mode of administration of medications (e.g., b.i.d.). This category has no UMLS equivalent.
Symptoms are the UMLS type Sign or Symptom.
Tests correspond to the UMLS semantic types Laboratory Procedure, Diagnostic Procedure, Clinical Attribute, and Organism Attribute.
Results represent the UMLS semantic types Laboratory or Test Results, and Finding.
Practitioners match Biomedical Occupation or Discipline, and Professional or Occupational Group.
Our corpus was annotated by two annotators based on the above definitions. Agreement, measured by the Kappa statistic, between the two annotators was 93% (strong inter-annotator agreement). The instances that the annotators did not agree on were reviewed and relabeled as necessary to generate a single annotated gold standard corpus. The resulting distribution of the semantic categories is in Table 1.1
Table 1.
Number of words in semantic categories.
| Category | Instances | Category | Instances |
|---|---|---|---|
| None | 27,228 | Diseases | 2,757 |
| Treatments | 2,685 | Symptoms | 1,410 |
| Results | 5,059 | Tests | 3,185 |
| Substances | 11 | Practitioners | 715 |
| Dosages | 3,086 |
Baseline Approach
Many systems implement a form of UMLS lookup to identify the key concepts in patient records. For evaluation purposes, we developed one such rule-based baseline system that maps noun phrases to UMLS semantic types. Like Long’s4, this system aims to find the longest string within a noun phrase that includes the head of the noun phrase (assumed to be the right-most noun) and that maps to one of the eight semantic categories. Every word in the noun phrase is then assigned that semantic category. In the case of phrases that map to multiple UMLS semantic types, the baseline returns multiple semantic types. E.g., face mask belongs to the UMLS semantic type Finding which maps to results, and to Medical Device which maps to treatments. Table 2 shows the expected values of precision and recall (assuming a uniform distribution over the predicted semantic categories for each word) of the baseline.
Table 2.
Precision (P), recall (R), and F-measure (F) for the baseline system.
| Class | P | R | F |
|---|---|---|---|
| None | 0.828 | 0.883 | 0.855 |
| Disease | 0.656 | 0.707 | 0.680 |
| Treatment | 0.548 | 0.726 | 0.625 |
| Test | 0.764 | 0.560 | 0.646 |
| Results | 0.404 | 0.358 | 0.380 |
| Dosages | 0.901 | 0.597 | 0.718 |
| Symptoms | 0.653 | 0.334 | 0.442 |
| Practitioners | 0.486 | 0.733 | 0.584 |
| Substances | 0.685 | 0.128 | 0.215 |
The baseline's F-measures on diseases, treatments, and tests are above 60%, confirming that mapping text to UMLS is an effective means of identifying these semantic categories. However, the F-measures for symptoms and results are discouragingly low; these categories often contain long, complex phrases and clauses that do not occur in UMLS. For example, consider the sentence: “A portable chest x-ray dated 9/5/2001 showed the [nasogastric tube to be in good position with a left sided PICC line at the superior vena cava]” where the results phrase is in square brackets. Using UMLS, the baseline recognizes nasogastric tube and left-sided PICC line individually as treatments, but fails to find the complete results phrase.
For practitioners and substances, the majority of errors are due to a mismatch between our definitions of these categories and the corresponding UMLS semantic types. The rest of the errors appear in determining the boundaries of semantic categories or are due to lack of useful context information.
Theoretically, the baseline could be improved through the addition of more rules. However, these rules would have to be manually adjusted to different corpora. Instead, we can use a statistical system based on orthographic, lexical, and syntactic features to automatically acquire the necessary rules that characterize the different semantic categories.
Comparison of Baseline with MetaMap
Before building our statistical SCR, we compared the baseline with MetaMap2, a system designed to map free text to concepts in the UMLS Metathesaurus. Table 3 shows that MetaMap outperforms the baseline only on diseases. It performs as well or worse than the baseline on all other categories.
Table 3.
Evaluation of MetaMap.
| Class | P | R | F |
|---|---|---|---|
| None | 0.715 | 0.922 | 0.805 |
| Disease | 0.742 | 0.667 | 0.806 |
| Treatment | 0.604 | 0.752 | 0.670 |
| Test | 0.577 | 0.282 | 0.461 |
| Results | 0.196 | 0.084 | 0.118 |
| Dosages | 0 | 0 | 0 |
| Symptoms | 0.727 | 0.321 | 0.445 |
| Practitioners | 0.411 | 0.195 | 0.265 |
| Substances | 0.269 | 0.138 | 0.182 |
MetaMap scores and returns multiple matches per phrase. We evaluated it based on the highest ranked UMLS categories it returned (several categories can tie for the top score) and computed the expected precision, recall, and F-measures assuming a uniform distribution over the predicted categories.
MetaMap gives poor performance on this data set because some categories, e.g., practitioners, contain proper nouns that cannot be found in UMLS; because many tests include numbers, which are missed; because the titles of medical professionals such as Dr. and M.D. get marked as diseases or tests; because elements of categories such as findings and results contain long, complex phrases and clauses only parts of which can be recognized by MetaMap; and because of ambiguities and definition mismatches between categories.
Statistical Semantic Category Recognizer
We use SVMs to predict the semantic category of each word (excluding punctuation) in the discharge summaries. If a word is predicted to belong to none of the semantic categories, then we label it as none.
We use the multi-class SVM implementation of LibSVM7 which combines several one-versus-one (rather than one-versus-all) classifiers. We apply this classifier to features that capture as much of the context and the characteristics of the target word (word to be classified) as possible. These features provide a very high dimensional space which makes SVMs particularly suited to our task: SVMs are robust to errors introduced by noisy features typically present in high dimensional spaces and effectively model the various semantic categories despite the curse of dimensionality. To minimize the risk of overfitting, we use linear kernels.
In this corpus, many words are associated with only one semantic category. Therefore, we use the target word itself as a feature. We capture the characteristics and the context of the target through orthographic, lexical, syntactic, and ontological (semantic) features.
Orthographic
Our orthographic features mark whether the target is capitalized or uppercase, and whether it contains numbers or punctuation. In our data, some treatments are capitalized; some tests appear in uppercase; many dosages and results contain numbers; and dates contain punctuation, but results don’t.
Lexical
Lexical features include:
The left and right lexical bigrams (uninterrupted strings of two words) of the target. Some bigrams are strongly correlated with a single category. For example, the left bigram status post is a strong indicator of treatments.
The heading of the section in which the target word appears. Discharge summaries contain sections such as Family History.
Syntactic
Medical discharge summaries contain many ungrammatical and fragmented sentences. Many syntactic parsers fail on such sentences. To obtain syntactic information under these conditions, we use the Link Grammar Parser8. This parser provides partial parses for those sentences that cannot be fully parsed. The partial parses contain important local syntax information that can be useful for SCR. For example, Link Grammar parses “John smokes two packs daily.”2 as follows.
From this parse, we extract syntactic bigrams for each word: a syntactic bigram consists of all words (and associated link labels) that are at a distance of at most two links. E.g., for smokes, its immediate left connection (a connection is a pair consisting of the link name and the word linked to) is {(John, Ss)}, marking the singular subject (Ss) of smokes. We represent the left syntactic unigrams of smokes with this set of connections. For each element of the left unigram set thus extracted, we find all of its immediate left connections, i.e., {(LEFT_WALL, Wd)}—LEFT_WALL represents the beginning of the sentence and Wd links the LEFT_WALL to the subject of the declarative sentence. The left syntactic bigram of the word smokes is then {{(LEFT_WALL, Wd)},{(John, Ss)}}. For words with no left or right links, we create syntactic bigrams using the two words immediately surrounding them with a link value of NONE.
For SCR, we used three types of syntactic features:
Syntactic bigrams of the target (referred to as syntactic bigrams). We hypothesize that certain semantic categories are characterized by their syntactic context. For example, the practitioners category is frequently the subject of the verb felt, as in, “The doctor felt that the patient was stable”. Studying the lexical bigrams around words would be sufficient to relate doctor and felt in this example; however, if modifiers are introduced, “The doctor, who came in yesterday, felt that the patient was stable”, deeper syntactic parsing is required to link doctor with felt.
The head of the noun phrase containing the target and the syntactic bigrams of the head (referred to as head bigrams)3. We hypothesize that links to the head of a noun phrase are more informative for SCR than links to the modifiers.
The part of speech of the target and of the words within a +/− 2 context window of the target.
Ontological
We used the UMLS predictions of the baseline as an input to the statistical system. We hypothesize that the UMLS semantic types will contribute to the performance for the categories that are well represented in UMLS.
Results and Discussion
We evaluated our system using tenfold cross-validation and computed F-measures for the complete feature set (i.e., all features), lexical features alone (i.e., lexical bigrams and section headings only), syntactic features alone (i.e., syntactic and head bigrams only), UMLS predictions from the baseline alone, and the target alone.
The results show that when run with all features, the statistical recognizer outperforms the baseline for all categories (the differences between the F-measures presented in Table 2 and Table 4 are significant at α = 0.05). The performance gain comes from instances such as the following:
Table 4.
Results when using all of the features.
| Class | Features | P | R | F |
|---|---|---|---|---|
| None | All features | 0.938 | 0.962 | 0.950 |
| Diseases | 0.911 | 0.899 | 0.905 | |
| Treatments | 0.924 | 0.901 | 0.912 | |
| Tests | 0.931 | 0.913 | 0.922 | |
| Results | 0.857 | 0.809 | 0.832 | |
| Dosages | 0.966 | 0.941 | 0.954 | |
| Symptoms | 0.901 | 0.815 | 0.856 | |
| Practitioners | 0.978 | 0.934 | 0.956 | |
| Substances | 0.934 | 0.853 | 0.892 |
The baseline marks the modifier resultant as a disease in the sentence, “However, the patient had a resultant [mediastinal hematoma]...” where the actual disease is shown in square brackets.
The baseline marks rehabilitation as a treatment instead of a facility in “Dr. Joe will see the patient at rehabilitation.”
Despite correctly identifying the semantic categories in the above cases, the statistical SC recognizer is not perfect. For example, it incorrectly maps infectious disease to diseases instead of practitioners in the sentence, “The patient was continued on Levaquin po antibiotics only per infectious disease recommendations”. Statistical approaches, in general, are susceptible to paucity of data. Therefore, words that occur infrequently get misclassified. Also, the results and symptoms categories still pose problems (despite the significant improvement, they have the lowest F-measures).
We investigated the contribution of syntactic, lexical, target word, and UMLS-prediction features to SCR in detail. We found that UMLS-prediction features are the least informative among the four feature groups (marked by the lowest F-measures in all categories in Table 5 and 6). The statistical SC recognizer with the UMLS-predictions gives very similar performance to the baseline; only the F-measures for treatments, tests, and results improve significantly. This is because the statistical approach learns the correlation between its input features and output class. If the correlation is weak, as it is for results and symptoms, then the errors propagate to the final classification. The observed improvement is due to elimination of irrelevant classes from the instances where the baseline system returned multiple categories.
Table 5.
Results when classifying based on target words alone and ontological features alone.
| Class | Features | P | R | F |
|---|---|---|---|---|
| None | Target alone | 0.856 | 0.957 | 0.904 |
| Diseases | 0.785 | 0.745 | 0.764 | |
| Treatments | 0.884 | 0.744 | 0.808 | |
| Tests | 0.815 | 0.847 | 0.831 | |
| Results | 0.732 | 0.467 | 0.570 | |
| Dosages | 0.827 | 0.758 | 0.791 | |
| Symptoms | 0.876 | 0.590 | 0.705 | |
| Practitioners | 0.874 | 0.622 | 0.727 | |
| Substances | 0.829 | 0.793 | 0.811 | |
| None | UMLS predictions alone | 0.825 | 0.891 | 0.857 |
| Diseases | 0.666 | 0.718 | 0.691 | |
| Treatments | 0.554 | 0.810 | 0.658 | |
| Tests | 0.806 | 0.649 | 0.719 | |
| Results | 0.477 | 0.343 | 0.400 | |
| Dosages | 0.903 | 0.597 | 0.719 | |
| Symptoms | 0.648 | 0.344 | 0.449 | |
| Practitioners | 0.545 | 0.701 | 0.613 | |
| Substances | 0.719 | 0.198 | 0.311 |
Table 6.
Results when classifying based on lexical features alone and syntactic features alone.
| Class | Features | P | R | F |
|---|---|---|---|---|
| None | Lexical alone | 0.844 | 0.931 | 0.886 |
| Diseases | 0.739 | 0.669 | 0.702 | |
| Treatments | 0.771 | 0.601 | 0.676 | |
| Tests | 0.812 | 0.684 | 0.743 | |
| Results | 0.751 | 0.648 | 0.696 | |
| Dosages | 0.929 | 0.884 | 0.906 | |
| Symptoms | 0.750 | 0.522 | 0.616 | |
| Practitioners | 0.889 | 0.727 | 0.800 | |
| Substances | 0.825 | 0.569 | 0.673 | |
| None | Syntactic alone | 0.901 | 0.939 | 0.919 |
| Diseases | 0.765 | 0.746 | 0.755 | |
| Treatments | 0.822 | 0.769 | 0.795 | |
| Tests | 0.831 | 0.803 | 0.817 | |
| Results | 0.796 | 0.733 | 0.763 | |
| Dosages | 0.931 | 0.898 | 0.914 | |
| Symptoms | 0.747 | 0.621 | 0.679 | |
| Practitioners | 0.911 | 0.890 | 0.900 | |
| Substances | 0.849 | 0.629 | 0.723 |
The targets themselves provide useful information for diseases, treatments, tests, and substances; in fact, the targets are more useful than lexical context for these categories (the F-measures are significantly higher than the corresponding values for lexical features, but not significantly higher than the values for syntactic features.) This is because the same actual diseases, treatments, tests, and substances repeat throughout the text; e.g., of the 33 occurrences of alcohol, 28 are substances. However, for more complex categories such as results and symptoms, or for categories involving numbers that do not repeat throughout the text, e.g., dosages, contextual features (lexical and syntactic) provide more information. E.g., the member of the practitioners category in square brackets in “[Infectious disease] was consulted.” is misclassified by the target-word-only model as a member of diseases but is correctly classified by the use of syntactic context. Syntactic context exposes the fact that disease is the subject of was consulted, which is a strong indicator of the practitioners category.
Finally, we notice that syntactic features outperform lexical features (all the F-measure increases are significant except for the increases in dosages and symptoms categories). To identify the most informative syntactic feature, we ran another set of experiments with lexical bigrams alone (i.e., the most useful lexical feature), syntactic bigrams alone, and head bigrams alone. We found that each of the tested features contributes differently to classification of each of the categories and therefore there is no clear winner. The bigrams that contribute the most to recognition of each category are shown in Table 7.
Table 7.
Best performing feature for each category.
| Category | Feature |
|---|---|
| None | Syntactic bigrams |
| Diseases | Head bigrams |
| Treatments | Syntactic bigrams |
| Tests | Syntactic bigrams |
| Results | Syntactic bigrams |
| Dosages | Lexical bigrams |
| Symptoms | Head bigrams |
| Practitioners | Head bigrams |
| Substances | Syntactic bigrams |
Note that in all but one case, syntactic bigrams outperform lexical bigrams. What is more, the syntactic features have different strengths and perform well when combined. E.g., in the sentence “At that time the [stroke fellow] was contacted.” the head bigrams correctly mark stroke fellow. However, the head bigrams have the disadvantage of associating the same features with all of the words in a phrase; this eliminates the distinction between heads and their modifiers and may lead to misclassification. Syntactic links associated with each word help maintain the distinction between modifiers and heads, preventing these mistakes.
Conclusion
We have described a statistical SC recognizer for discharge summaries. We have shown that for clinical text, contextual clues (lexical and syntactic) provide stronger indications of semantic categories than information extracted from UMLS. We have also described a method for using the output of the Link Grammar parser to capture the syntactic context of words, and have shown that these syntactic contextual clues are the strongest determinants of certain semantic categories.
Acknowledgments
Acknowledgements
This research was funded in part by the National Institutes of Health through grant number R01-EB001659 from the National Institute of Biomedical Imaging and Bioenginering; grant number N01-LM-3-3513 on National Multi-Protocol Ensemble for Self-Scaling Systems for Health from National Library of Medicine; and grant number U54-LM008748 on Informatics for Integrating Biology to the Bedside from National Library of Medicine.
Footnotes
The numbers indicate the total number of words tagged as the corresponding semantic category. For example, the phrase coronary artery disease yields three disease instances.
Op links verbs to their plural objects; Dmc links determiners to their plural nouns; MVa connects verbs to their modifiers; Xp links periods to words.
For words not occurring in noun phrases, this feature is identical to the target's syntactic bigrams.
Contributor Information
Tian He, MIT CSAIL, Cambridge, MA 02139
Peter Szolovits, MIT CSAIL, Cambridge, MA 02139
Ozlem Uzuner, State University of New York, Albany, NY 12222
References
- 1.Friedman C, Alderson P, Austin J, Cimino J, Johnson S. A general natural language text processor for clinical radiology. JAMIA. 1:1994. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson A. Effective mapping of biomedical text to the UMLS Metathesaurus: the Metamap program. AMIA. 2001 [PMC free article] [PubMed] [Google Scholar]
- 3.Delbecque T, Jacquemart P, Zweigenbaum P. Indexing UMLS semantic types for medical question-answering. Studies in Health Technology and Informatics. 2005;116 [PubMed] [Google Scholar]
- 4.Long W. Extracting diagnoses from discharge summaries. AMIA. 2005 [PMC free article] [PubMed] [Google Scholar]
- 5.S. Soderland, D. Aronow, D. Fisher, J. Aseltine, and W. Lehnert, Machine learning of text analysis rules for clinical records, Technical Report: UMASS at Amherst. Center for Intelligent Information Retrieval, 1995.
- 6.M. Averbuch, T. Karson, B. Ben-Ami, O. Maimon, and L. Rokach, Context-sensitive medical information retrieval. MedInfo, 2004. [PubMed]
- 7.C. Chang and C. Lin, Libsvm: a library for support vector machines, 2001.
- 8.D. Sleator and D. Temperley, Parsing English with a link grammar, Carnegie Mellon University Computer Science Technical Report CMU-CS-91-196, 1991.