Abstract
A substantial effort of the biological community involves the development of model organism databases containing key genomic information concerning specific organisms. This paper describes a developing natural language processing (NLP) tool, which is aimed at assisting curators of the Mouse Tumor Biology (MTB) Database of the Mouse Genome Informatics (MGI) group by helping them quickly find key information in the articles.
Introduction
Model organism databases, which contain key genomic information concerning specific organisms, provide significant support for ongoing research. For example, MGI consists of a comprehensive database of laboratory mouse, which is very significant to the biological research community. For those data annotated manually, curators locate the information by: 1) finding the appropriate journal articles, 2) identifying the relevant information by reading the articles, and 3) entering the information into a database using an appropriate coding-system. A tremendous amount of new biological discoveries are being continually reported in the literature, making it virtually impossible for curators to keep pace with the volume. Therefore, an automated tool that would assist curators to quickly find relevant articles and key information in the articles would be a valuable tool that should increase their productivity. Natural language processing (NLP) is one of the technologies that could potentially be used for this purpose. Within the past few years there has been an increased effort to develop NLP systems that extract and acquire biological knowledge from the literature. However it is not clear how to integrate an NLP system into the workflow of the database curation process or what the effect of such a system would actually be. The project we describe here concerns development of an online tool for MGI curators. This tool utilizes an existing NLP system called BioMedLEE [1] (Biology & Medical Language Extracting and Encoding), which is based on another NLP system: MedLEE (Medical Language Extraction and Encoding), which captures and encodes medical information from patient reports. The major functionality of the NLP-based curator tool is to summarize key information in an article based on the curators’ needs, and to highlight the information within the article.
Methods
For this project we are collaborating with the MTB group of MGI, which collects mouse tumor findings from articles [2]. This project involved the following steps: 1) working with the curators to determine which information is useful to them, 2) determining the appropriate presentation of the information, such as determining the terms to display and their order, 3) developing an effective highlighting function that links the extracted data to the original text to allow easy validation of the captured information, and 4) development of an online interface prototype, which was implemented with Java and a MySQL database, which contained the output structured by BioMedLEE.
Results
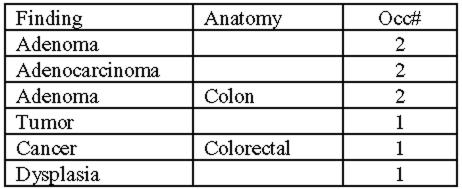
We used this tool to extract and display findings from 25 abstracts that were previously manually processed by MTB curators, and compared our results to theirs. Fig. 1 shows tumor findings obtained by the NLP tool from one. It has three columns: tumor terms from the original abstract, tissues of the tumor, and the number of occurrence of the tumor findings. The original abstract is displayed side by side with this finding list. After clicking on a finding, the corresponding terms in the original abstract will be highlighted so that curators can easily verify the extracted finding. Feedback from curators who reviewed the extracted findings was positive. Further evaluation will be performed to determine the performance and the effectiveness for curation.
Fig 1.
Displayed findings of an abstract by NLP tool
Conclusion
This paper describes a prototypical NLP tool for curators of model organism databases. It aimed to represent their information needs and increase their productivity, and was developed with curators from MGI. Although this tool is promising, the effectiveness and the user satisfaction will have to be evaluated in the next step of this project.
Acknowledgments
This work was supported by grants LM07659/LM008635 from the NLM, HG00330 from the NHGRI, and CA89713 from the NCI
References
- 1.Friedman C, Chen L. Extracting Phenotypic Information from the Literature via Natural Language Processing. Medinfo. 2004;2004:758–62. [PubMed] [Google Scholar]
- 2.Näf D, Krupke DM, Sundberg JP, Eppig JT, Bult CJ. The mouse tumor biology database: a public resource for cancer genetics and pathology of the mouse. Cancer Res. 2002;62(5):1235–40. [PubMed] [Google Scholar]