Abstract
Through Natural Language Processing (NLP) techniques, information can be extracted from clinical narratives for a variety of applications (e.g., patient management). While the complex and nested output of NLP systems can be expressed in standard formats, such as the eXtensible Markup Language (XML), these representations may not be directly suitable for certain end-users or applications. The availability of a ‘tabular’ format that simplifies the content and structure of NLP output may facilitate the dissemination and use by users who are more familiar with common spreadsheet, database, or statistical tools. In this paper, we describe the knowledge-based design of a tabular representation for NLP output and development of a transformation program for the structured output of MedLEE, an NLP system at our institution. Through an evaluation, we found that the simplified tabular format is comparable to existing more complex NLP formats in effectiveness for identifying clinical conditions in narrative reports.
INTRODUCTION
Clinical narratives are a significant component of the Electronic Medical Record (EMR). The text in these documents (e.g., radiology reports, cardiology reports, and discharge summaries) include a wealth of information about clinical findings in patients. One challenge is extracting these findings to assist with applications such as patient management, decision support, quality assurance, and clinical research. In response to this, Natural Language Processing (NLP) systems have been developed to identify, extract, and encode information within clinical narrative text. Standards, such as the eXtensible Markup Language (XML), offer a solution for exchanging and sharing complex and nested output from NLP systems. While such formats are useful for subsequent automated applications, they may not be suitable for presentation of information to and direct use by end-users such as clinicians and clinical researchers.
To address this issue, we are exploring the transformation of the nested structured output from an NLP system into a tabular format, which may be more suitable and usable for particular applications. This simplified format could facilitate review of the natural language processed clinical narratives and would allow users to import the output into popular programs (e.g., spreadsheet and database programs like Microsoft Excel and Access) for further analysis. In this paper, we present the design of a 21-field tabular representation for NLP output based on analysis of XML output from the Medical Extraction and Encoding (MedLEE)1–3 system at NewYork-Presbyterian Hospital (NYPH). The goal for creating this format was to simplify the complexity and nesting of NLP output while minimizing information loss. Through an evaluation, we found that the tabular format is comparable to existing NLP formats, thus providing an alternative representation of natural language processed clinical narratives that may be preferred by certain end-users or applications.
BACKGROUND
Current NLP tools are primarily focused on producing structured output geared for use by automated applications rather than dissemination and direct use by clinicians and researchers. MetaMap, produced by the National Library of Medicine, was designed to discover UMLS Metathesaurus concepts in biomedical text4. The output of MetaMap includes a list of candidates along with scores representing the strength of the candidate mapping and is available in two formats: ‘machine output’ and ‘fielded output’ (that produces mutli-line tab-delimited output) for further processing. At the University of Utah, an event-based model was created to represent medical information for NLP and other applications5. In this model, frames are used to represent medical events where slots are name-value pairs for each attribute. In a recent study, MPLUS (their latest Natural Language Understanding application) and MetaMap were used to convert clinical documents into XML-based Clinical Document Architecture (CDA) versions for automated generation of medical problem lists6.
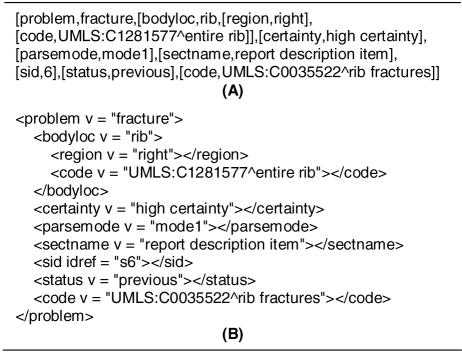
MedLEE is a natural language processing system at NYPH that has been used to extract and encode information in clinical narratives for a number of applications and studies1–3. Originally developed to handle chest radiograph reports, this system has been extended over the years to support cardiology reports, pathology reports, discharge summaries, and all of radiology. For each report, MedLEE produces a set of primary findings (e.g., problem, procedure, device, and medication) along with associated modifiers (e.g., certainty, change, body location, and frequency) where the findings can be structured in a variety of formats such as XML. This output is based on frames in the form Type-Value-Modifiers where Type and Value refer to the primary finding followed by Modifiers, which are also frames following the same format, thereby allowing for nesting of modifiers. For example, in the sentence “Old fractures of the right ribs are noted”, the value of the primary finding is fracture with type problem. Modifiers for this finding include body location (bodyloc), region, certainty, and status with the values rib, right, high certainty, and previous, respectively. UMLS codes may be available for the primary finding as well as certain modifiers (e.g., bodyloc), and is represented as an additional modifier called code (e.g., ‘C0035522’ corresponding to ‘rib fractures’ is assigned to this finding). Figure 1 depicts this example sentence in the line and XML representations of MedLEE, both of which are equivalent. Several studies have explored developing different representations for MedLEE output to determine their effect on performance of machine learning algorithms and to create different views of the patient record7–10.
Figure 1. MedLEE Output Formats.
Output (simplified for demonstration purposes) for the sentence “Old fractures of the right ribs are noted” in line (A) and XML (B) formats.
METHODS & RESULTS
Overview
As a first attempt towards simplification and dissemination of NLP system output, we sought to create a tabular format to represent the information produced from applying an NLP system to clinical narrative reports that minimizes loss of information and relations (i.e., information about a finding as qualified by modifiers). The first step was to analyze a set of reports to identify the typical content and structure for determining which information to flatten or filter. This analysis involved studying the XML structured output of MedLEE for radiology reports, specifically chest radiographs, as the output is complex and often consists of considerable nesting. After this analysis, we designed a new structure for organizing each finding and its associated modifiers into individual lines. With this structure defined, we developed a program to transform MedLEE XML output into the tabular representation. To evaluate the efficiency of this new condensed structure and the affect on subsequent applications, we compared results of queries for clinical conditions to those from a previous published study.
Analyzing Report Content and Structure
MedLEE XML output for chest radiograph reports from 2000 through 2004 were obtained and analyzed. All XML elements (representing the type of the primary finding or modifier) and ‘v’ attributes (representing the value of the primary finding or modifier) were extracted. For each unique element and attribute, frequency counts were calculated to assess the semantic content of these reports. In addition, we looked at a subset of the reports to study the structured output with respect to repeated and nested modifiers.
Based on analysis of the set of over 500,000 reports, we found that the more frequent elements included the primary finding type (e.g., problem and procedure) and the following modifiers: bodyloc, certainty, code, change, descriptor, parse mode (parsemode), region, section name (sectname), and status. From the subset of reports, repeated modifiers were found to include certainty, change, degree, descriptor, quantity, region, and status. Finally, nesting was found to occur for a number of modifiers and most significantly for bodyloc where nested modifiers included region, locative, and additional bodyloc. Table 1 contains examples of repeated modifiers and nested modifiers for body location.
Table 1.
Repeated and Nested Modifiers.
| Modifier Type | Example Sentence |
|---|---|
| status (repeated) | Previously suspected pulmonary edema has resolved |
| descriptor (repeated) | Freely layeringsmall right pleural effusion |
| bodyloc (nesting) | Question lesionbehindrightfirstcostocartilage |
primary finding (italics), bodyloc (bold italics), repeated modifiers (underlined), nested locative and region modifiers for bodyloc (dotted line)
Designing Tabular Representation
Based on the aforementioned analysis of chest radiograph reports and expert knowledge about MedLEE, we created a representation to condense the information into a tabular structure consisting of 21 fields. In order to reduce the amount of information represented in this structure while preserving key information, rules were created for merging findings and modifiers to handle repeats and nesting. Table 2 defines the fields and several rules for the tabular representation.
Table 2.
Fields and Rules for Tabular Representation.
| Field | Description and Rules (in italics) |
|---|---|
| 1. mrn |
|
| 2. date |
|
| 3. acc_no |
|
| 4. report_type |
|
| 5. finding_id |
|
| 6. primary_type |
|
| 7. primary_value |
|
| 8. primary_code |
|
| 9. bodyloc |
|
| 10. bodyloc_code |
|
| 11. region |
|
| 12. laterality |
|
| 13. change |
|
| 14. descriptor |
|
| 15. problemdescr |
|
| 16. certainty |
|
| 17. status |
|
| 18. other_mod |
|
| 19. sectname |
|
| 20. sid |
|
| 21. parsemode |
|
With the exception of the body location modifier, any other modifiers with repeat or multiple values were concatenated since they are infrequently accessed. For example, the output for the sentence “Right-sided central venous catheter is seen with the distal tip at the junction of the SVC/right atrium” includes four region modifiers: central, tip, distal, and junction. With the tabular format, this would be represented in the region field as ‘central^tip^distal^junction’. For the other_mod field, ‘:’, ‘^’, and ‘;’ are used to separate the different modifier types and values (e.g., ‘degree:low degree^partial degree;locative:behind’). Due to the complexity and fine granularity of nesting, decisions were made about which nested modifiers to include and exclude at this time. For example, in the case of bodyloc, nested bodyloc are listed in separate lines with all other nested modifiers concatenated in the respective fields. However, in other cases, such as change (e.g., ‘increase’), only nested certainty (e.g., ‘no’) values are kept and concatenated (e.g., ‘increase – no’) while for others like position, all nested modifiers are ignored because we did not find that such a fine level of granularity was necessary.
Transforming XML Output
Once the tabular representation was defined, we developed a program using eXtensible Stylesheet Language Transformations (XSLT) and Perl to convert MedLEE XML output. XSLT is a technology for transforming XML documents into other useful forms11. The XSLT processor takes an XSLT stylesheet consisting of template rules for transforming input XML documents into new XML or other formats (e.g., HTML, delimited files, or plain text). We created an XSLT stylesheet to support and transform the features of MedLEE XML into a format for further processing by Perl. The resulting Perl script was responsible for reading in the MedLEE XML documents, applying the stylesheet using the XML::LibXSLT and XML::LibXML modules, and converting the XSLT output into a 21-field pipe-delimited format. In order to reduce the data, this script also replaced values in some fields with abbreviated versions. For example, ‘report description item’ was substituted with ‘description’ in the sectname field and the parsemode ‘mode1’ was reduced to ‘1’. Figure 2 presents the tabular output for several example sentences from chest radiograph reports.
Figure 2. MedLEE Output in Pipe-Delimited Tabular Format.
These three examples highlight how findings with multiple region values (A), findings with several modifiers in the other_mod field (B), and findings with multiple body location modifiers (C) are represented in the 21-field tabular format.
Evaluating Effectiveness of the Tabular Format
To assess the efficiency of the tabular representation, we simulated a study published in 200212. This study evaluated the translation of clinical information in chest radiograph reports by MedLEE to determine the frequency of 24 clinical conditions in reports spanning a ten-year period from 1989 to 1998 (n = 889,921). Queries for identifying conditions in specific sections of the reports (e.g., ‘neoplasm’ in the description or impression section and ‘sputum’ in the clinical information section) were written as a set of Perl scripts and were run on the line output format of MedLEE (shown in Figure 1A). The results from these queries were stored and accessible for comparison to results from the current study.
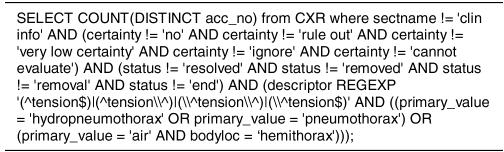
Out of the 889,921 reports, we were able to retrieve 99.9% of the reports that had been reprocessed by the latest version of MedLEE and stored in XML format. We applied the Perl transformation script to this set of reports, which produced a tabular file consisting of over 13 million lines. For this study, we imported the tabular file into a MySQL database and rewrote the queries for identifying the 24 clinical conditions in Structured Query Language (SQL). Figure 3 presents one query for ‘tension pneumothorax’. Each SQL query was run and refined as needed and results compared to those from the 2002 study. Although the number of reports returned differed across the conditions, we were able to match from 87.02% to 100.00% (mean 96.15% ± 3.39%) of the reports. We found that in some cases the 2002 query results consisted of reports missed by our study and vice versa. Table 3 presents the breakdown for a subset of the queries. Using the unique accession numbers, we manually referred back to the original reports identified by each query for both studies to determine the reason for exclusion from the respective study. Based on analysis of a sample of reports unique to each study, we categorized the reasons for the differences.
Figure 3. Query for Tension Pneumothorax.
MySQL query for finding the clinical condition in the set of reports.
Table 3.
Results of Queries for Subset of Clinical Conditions.
| Query | TP | FN | FP |
|---|---|---|---|
| Sputum (clinical information section) | 2,186 (100.0%) | 0 | 651 |
| Rule out rib fracture (clinical information section) | 2,883 (92.32%) | 240 | 101 |
| Tension pneumothorax (description/impression section) | 369 (95.60%) | 17 | 40 |
| Neoplasm (description/impression section) | 40,731 (89.65%) | 4,703 | 6,419 |
TP: True Positives; reports found by both studies (% of 2002 reports)
FN: False Negatives; reports detected by the 2002 study and not the current study
FP: False Positives; reports detected by this study and not the 2002 study
Using the 2002 results as a reference, reports missed by our current study (‘false negatives’) were due to modifications in the MedLEE system since the original study, specifically changes in the lexicon and output form, and were not due to the tabular format. While we accounted for some lexical changes during initial query refinement (e.g., the query for determining frequency of bullet and stab wounds was extended to include the finding ‘bullet fragment’), other variations were not discovered until later. Changes in the output form included use of new or different modifiers to qualify primary findings (e.g., use of the descriptor problemdescr for nested problems). Modifications to the queries to take into account these lexical and output form changes would reduce the false negatives. In looking at reports unique to our study (‘false positives’), we confirmed that some were indeed true positives and were correctly included while others were incorrectly included due to concatenated values (i.e., modifier types and values delimited ‘:’, ‘^’, and ‘;’). Further refinement of the queries to perform pattern matching rather than exact matches would eliminate these false positives.
DISCUSSION
Our motivation for the development of a tabular representation of output from NLP systems, such as MedLEE, was to create an easy-to-use simplified structure while maintaining essential information about clinical findings (as represented by modifiers). By providing a format that can be imported into programs commonly used by clinicians and clinical researchers, we may be able to facilitate their use of clinical information in narrative reports for improving patient management and other healthcare functions. Other users (e.g., biomedical researchers and biostatisticians) could also benefit from a tabular representation for purposes including data mining, information retrieval, and statistical analyses.
To guide the design of the tabular representation, we first obtained descriptive statistics for a large set of chest radiograph reports to determine frequently and infrequently occurring modifiers. Based on the analysis, we created a structure to reduce the complexity of information and handle nesting of modifiers. Through an evaluation, we found that the 21-field tabular format is able to represent the content and structure of natural language processed clinical narratives in a condensed form while remaining effective for identifying a range of conditions. The results of queries for clinical conditions in the evaluation were comparable to a more complex NLP format and differences could be attributed to changes to components of MedLEE and not the tabular form. We found that the tabular structure could be easily loaded into a database program and queried using SQL. On the other hand, some disadvantages of this format include loss of linkages to the original narrative text in reports (available in the XML format), loss of some fine-grained information, and the complexity of queries required to process the less frequently occurring concatenated values in fields. Next steps include performing comprehensive evaluations of the content, structure, and usability of the tabular representation for NLP output.
While the tabular structure seeks to condense and simplify NLP output, additional techniques may be warranted for further reducing the amount of information and simplifying retrieval. The transformation of ten years of chest radiograph reports in this study produced a file containing over 1 GB of data and 13 million lines (compared to an estimated 8 GB and 116 million lines for the XML output). This amount of information could be difficult to load into certain programs and reviewed by users. Filtering techniques can be applied to include only the most valuable clinical information and exclude unneeded information. For example, inclusion and exclusion criteria could be defined and applied to the certainty and status fields to filter out findings with low certainty, negated findings, or past findings, which may not be of interest to clinicians. Additionally, techniques such as creating an index or merging rows can be used to compress the information. Other mechanisms may still be needed to assist users with tasks like searching for particular findings, which would require familiarity with the vocabulary or UMLS codes. To address this issue, supplementary information such as translation tables and predefined queries could be distributed.
In this work, we have begun to explore the use of XSLT to transform complex XML output generated by an NLP system. The Perl transformation script was applied to chest radiograph reports in this study, but could be used for other types of reports, such as pathology reports and discharge summaries. Other formats may be desired and customized stylesheets can be created to transform the XML into the most useful form for a given user or application.
NLP systems are playing an increasingly important role in the biomedical domain. We initially created the tabular format to represent complex and nested NLP output for the purpose of dissemination to clinicians and clinical researchers. Another potential application of this representation is as a standard for exchanging and sharing output produced by different NLP systems. The existence of a formal simplified representation for NLP output could facilitate the use and comparison of information captured in clinical and biomedical text, which may be represented at different levels of complexity and granularity by the respective system. Further work is needed to determine if this tabular format sufficiently represents the output from varying NLP systems or if modifications are needed to create a standard simplified representation that can support a range of systems for the purposes of sharing NLP output.
CONCLUSION
Natural language processing systems are a valuable resource for capturing and presenting information in clinical narratives. The ability to structure this information in a suitable format for common programs (e.g., spreadsheet or database) can facilitate use by different end-users for their specific needs. We have designed and implemented a tabular format for natural language processed clinical narratives and found that this representation was adequate for a variety of clinical queries. The availability of a format that simplifies NLP output may be valuable for dissemination to clinicians and researchers for tasks including patient management and clinical research.
Acknowledgments
This work is supported in part by grants LM007659, LM008635, and LM006910 from the National Library of Medicine.
References
- 1.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language. J Am Med Inform Assoc. 1999;6(1):76–87. doi: 10.1136/jamia.1999.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 5.Huff SM, Rocha RA, Bray BE, Warner HR, Haug PJ. An event model of medical information representation. J Am Med Inform Assoc. 1995;2(Mar–Apr;)(2):116–34. doi: 10.1136/jamia.1995.95261905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meystre S, Haug PJ. Automation of a problem list using natural language processing. BMC Med Inform Decis Mak. 2005;5:30. doi: 10.1186/1472-6947-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilcox A, Hripcsak G. Medical text representations for inductive learning. Proc AMIA Symp. 2000:923–7. [PMC free article] [PubMed] [Google Scholar]
- 8.Seol YH, Johnson SB, Starren J. Use of the Extensible Stylesheet Language (XSL) for medical data transformation. Proc AMIA Symp. 1999:142–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Friedman C. CliniViewer: a tool for viewing electronic medical records based on natural language processing and XML. Medinfo. 2004;11(Pt 1):639–43. [PubMed] [Google Scholar]
- 10.Krauthammer M, Hripcsak G. A knowledge model for the interpretation and visualization of NLP-parsed discharged summaries. Proc AMIA Symp. 2001:339–43. [PMC free article] [PubMed] [Google Scholar]
- 11.XSL Transformations (XSLT) Version 1.0. Available at: http://www.w3.org/TR/xslt.
- 12.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224(1):157–63. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]