Abstract
Drug information sources use named classes to assist in navigating and organizing information. Some of these classes describe drugs from multiple perspectives (e.g., both structure and function). The National Drug File – Reference Terminology (NDF-RT) is a drug information source that augments a “legacy” classification system via a formal reference model that groups drug classes into the following high-level categories: Chemical Structure, Cellular or Sub-Cellular Mechanism of Action, Organ- or System-Level Physiological Effect, and Therapeutic Intent.. We examined drug class names from three sources to better understand their information content and evaluate NDF-RT’s semantic coverage. On average, class names contain more than 1.5 attributes. NDF-RT’s categorical reference model accommodates more than 76% of the information identified in drug class names. A new NDF-RT reference axis of drug formulations could improve NDF-RT’s coverage to 85%. The distinction between Physiological Effect and Therapeutic Intent, prompted many questions among reviewers, suggesting that further clarification of these ideas is required. Careful review of existing classification schemes may guide structured terminology and ontology development efforts toward greater fidelity to deployed information sources.
Introduction
Grouping drugs into classes based on salient similarities helps users navigate and organize complex and rapidly-changing pharmaceutical information. For example, consider the package insert for Renese® tablets (a thiazide diuretic used in the treatment of edema and hypertension) as listed on the DailyMed Web site.1 The label itself identifies Renese® as a ‘thiazide,’ a ‘diuretic,’ and a ‘sulfonamide-derived drug.’ SNOMED CT2 lists polythiazide (the active ingredient in Renese®) as a ‘saluretic’ and a ‘thiazide diuretic,’ and includes it in the higher-level categories of ‘diuretic,’ ‘cardiovascular drug,’ and ‘renal drug.’ The Department of Veterans Affairs Veterans Health Administration (VHA) National Drug File (NDF) categorizes this ingredient under ‘diuretics/related preparations.’ The National Library of Medicine’s (NLM) Medical Subject Headings (MeSH)3 shows polythiazide as a ‘benzothiadiazine,’ an ‘anti-hypertensive agent,’ a ‘diuretic,’ and a ‘sodium chloride symporter inhibitor.’ MedLinePlus4 includes patient-oriented information on polythiazide under the heading ‘diuretics, thiazide (systemic).’ These many classifications provide substantial and varied information about this medicine’s structure, use, and mode of action.
The National Drug File – Reference Terminology (NDF-RT)5 is an ongoing project to extend VHA’s NDF. NDF is used today to order medications electronically in the VHA’s hospitals and clinics. NDF groups all orderable drug products into exactly one of 480 classes.* This single-class structure has obvious limitations: it is impossible to categorize a drug as both an “antihypertensive” and a “beta-Blocker.”
This study seeks to characterize the ways in which drug information sources classify drugs and determine the extent to which NDF-RT can represent this information.
Background
Classification schemes can be divided into those that require each member to fit into exactly one class (e.g., alphabetical, weight), and those that allow membership in multiple classes (e.g., ingredients, indications). In any case, a classification scheme should be complete (all possible members should fit into one or more class) and non-overlapping (the same information should be covered in only one class). Modern reference terminologies recognize that multiple classification schemes may be helpful to a diverse user community.
Drug classes themselves can be grouped into categories. For example, we can identify a comprehensive, non-overlapping set of chemical structure classes, treated diseases, targeted body systems, and so on. These “categories” of classes have no members in common (although of course the drugs thus organized belong in all of them).
Although they are frequently related, a drug’s membership in a class does not automatically connote membership in any other class. Structurally similar drugs treat different diseases, as in the case of trazodone, an antidepressive agent, and ketoconazole, an antifungal agent, which are both ‘piperazines’ according to MeSH. Similarly, drugs with different modes of action can treat the same disease, as in the case of ranitidine, a histamine antagonist, and misoprostol, a stomach lining protector, both used to treat stomach ulcers.
Deployed drug information classification systems combine classes from disjoint categories into a single system, and even into a single class. For example, the NDF class non-steroidal anti-inflammatory analgesics’ describes three separate drug attributes: drugs in this class do not contain steroids, do reduce inflammation, and also relieve pain. The MedLinePlus category narcotic analgesics for surgery and obstetrics (systemic)’ is even more complex.
Just as reference terminologies serve multiple user communities via multiple navigation paths, computerized decision support applications need to perform reasoning tasks based on multiple criteria. An allergy to penicillin, for example, usually translates to a warning against prescribing drugs that are structurally similar to penicillin. Conversely, analyzing medication compliance among diabetics would be better served by a treatment-focused classification than a structural one. Just as it is easier for a clinician to navigate and remember a relatively smaller set of drug classes (as opposed to the thousands of available drug products), it is easier for knowledge engineers to build rules based on drug classes rather than on enumerated lists of individual products. Therefore, we assert that explicit relationships between drugs and orthogonal categories of fine-grained classes will empower the development and improve the maintainability of computer-based decision support tools.
NDF-RT supports a wide range of computer-based tasks, including ordering, documentation of care, decision support and interoperability with external systems. NDF-RT seeks to provide the computer-empowering benefits of a formal reference terminology (as defined elsewhere)6 while preserving VHA’s investment in NDF-compatible software and systems. To meet these goals, NDF-RT combines NDF’s hierarchical drug classification with a multi-categorical reference model. Following the Prodigy project,7 NDF-RT’s reference model includes a category of drug classes describing Chemical Structure similarities, cellular or sub-cellular Mechanism of Action, and tissue-, organ-, or body system-specific Physiological Effect. While Prodigy characterized the primary drug-disease relationship as “Indication,” NDF-RT chose the name Therapeutic Intent, indicating a practical distancing from the exacting and often verbose indications found in the FDA-approved package insert. NDF-RT is developed using Apelon, Inc.’s Terminology Development Environment8 (a description logic-enabled vocabulary creation software tool). The categorical axes named above are instantiated as separate, hierarchical sets of reference terms.
Methods
In addition to the drug categorizations already present in NDF-RT, an ad hoc analysis of several drug knowledge bases revealed two additional information types. These are information about the drug’s Formulation (including packaging, administration and regulatory status) and its Non-Patient Activities (as in the case of many anti-infective categories that describe the drug’s action in terms of an infectious organism). Finally, we included an Other column to capture classificatory information not covered by the other categories. Prompted by Cimino9 and Lau,10 we also noted Self-referential or “Not Elsewhere Classified” classes, i.e., classes that only make sense given an understanding of other classes.
We performed detailed analysis on NDF’s 480 classes, the 170 formulary classes developed for use in the new Medicare Part D benefit,11 and the 298 classes from a proprietary drug knowledge base. Several of the authors (BAB, SHB, PLE, MSE, DAF, STR, DLW) reviewed the classes using a spreadsheet similar to Figure 1. For each class name, the reviewer marked a cell if the class described a similarity of the listed type. Since our goal was an inventory of the ways in which drugs are classified and a determination of whether or not such a classification was covered by NDF-RT’s existing categories, we sought consensus among the reviewers. Because of this information-sharing process, the kappa statistic is not reported. Each class could be assigned zero or more aspects of similarity by each reviewer. For the prespecified categories (Chemical Structure, Mechanism of Action, Physiological Effect, Therapeutic Intent, Non-patient Action, and Combination Category), we included a result if two or more reviewers agreed that the category described the listed type of similarity. For the “Other” column, identification by any one reviewer was sufficient.
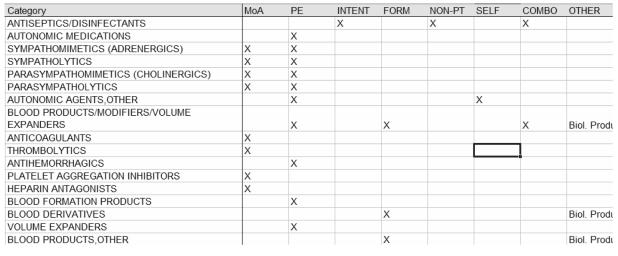
Figure 1. Selected drug categories from NDF, with categorization attributes identified by a reviewer.
MoA = Cellular or Subcellular Mechanism of Action, PE = Organ-, Tissue- or System-Specific Physiological Effect, FORM = Formulation, NON-PT = Non-host Activity, SELF = Self-Referential (relies on another category for definition), COMBO = Groups more than one kind of drug.
Results
As shown in Table 1, at least two reviewers agreed on a total of 976 separate descriptors in the 480 NDF drug classes, an average of 2.03 attributes per class. The 170 Medicare Part D classes revealed 249 attributes, an average of 1.46. The 298 classes from the commercial drug knowledge base yielded 461 (average 1.55).
Table 1.
Attributes by category from three drug classifications.
| Medicare | Comm. | ||
|---|---|---|---|
| NDF | Part D | KB | |
| Chemical Structure | 130 | 29 | 93 |
| Mechanism of Action | 89 | 35 | 51 |
| Physiological Effect | 192 | 37 | 109 |
| Therapeutic Intent | 318 | 88 | 123 |
| Formulation | 97 | 28 | 40 |
| Non-patient Activity | 47 | 27 | 27 |
| Other | 103 | 5 | 14 |
| Total | 976 | 249 | 461 |
| Number of Classes | 480 | 170 | 298 |
| Average Attributes per Class | 2.03 | 1.46 | 1.55 |
NDF = National Drug File, Comm. KB = Commercial Knowledge Base (existing NDF-RT categories in italics)
The relatively large number of “Other” attributes found in NDF stems primarily (76/103) from a set of class names describing investigational drugs. No analogous classes are found in the other two sources. Reviewers also used “Other” for biological products (e.g., blood products, vaccines) and for “generational” classes such as ‘1st generation cephalosporins.’
Table 2 shows the distribution of the number of classificatory attributes in the three drug information sources. Most class names contained either one or two attributes of similarity. Examples of single-attribute classes include ‘ACE inhibitors’ (mechanism of action), ‘salicylates’ (chemical structure) and ‘anti-emetics’ (therapeutic intent). Examples of two-attribute classes include ‘antihistamines, piperazine’ (mechanism of action and chemical structure) and ‘beta-blockers, topical ophthalmic’ (mechanism of action and formulation).
Table 2.
Distribution of Descriptors by Source.
| Medicare | Commercial | ||
|---|---|---|---|
| No. of Descriptors | NDF | Part D | KB |
| 0 | 0 | 10 | 6 |
| 1 | 119 | 88 | 160 |
| 2 | 238 | 56 | 101 |
| 3 | 111 | 15 | 27 |
| 4+ | 12 | 1 | 4 |
| Total Categories | 480 | 170 | 298 |
NDF = National Drug File, KB = Knowledge base.
Nearly 11% (103/948) of the classes in these three sources can only be understood in terms of other classes. These “not elsewhere classified” classes pose extra difficulties for computer-based decision support tools, since they provide no explicit clue to what content is included. This violation of one of Cimino’s desiderata for controlled vocabularies9 contrasts with Lau’s finding.10 Of the descriptive attributes found in the NDF classes, 74.69% (729/976) are from the category types already included in the NDF-RT reference model. For the Medicare Part D classes, the corresponding figure is 75.9% (189/249). NDF-RT’s categories cover 81.56% (376/461) of the commercial knowledge base class attributes.
The most frequently found category of information in all three sources is Therapeutic Intent, followed by Physiological Effect. For 16 of 948 (1.69%) of the classes, two or more reviewers did not agree on the intent of the class.
Discussion
Drug classification is ubiquitous. One notable recent example of the economic and clinical importance of drug classes is in the new Medicare Part D benefit, part of the Medicare Modernization Act of 2003. This law requires health plans to reimburse beneficiaries for at least two drugs in each of a specially constructed list of drug classes.12 More on-formulary classes means more complexity and more inventory for health plans and pharmacies, but also means more flexibility for doctors and patients. Fewer classes (therefore fewer drugs required to be included on the formulary) translates to fewer therapeutic options for beneficiaries and fewer economic opportunities for drug companies to make incremental improvements to existing drugs. The classification system may put the interests of drug companies, health plans and patients in direct conflict.
Previous evaluations of NDF-RT have described the methods for instantiating the reference model relationships,5 the coverage of the concepts in the reference hierarchies,13 and the extensibility of the model to novel domains.14 This is the first study to analyze NDF-RT’s multi-category reference model in terms of the legacy terminology it seeks to augment and the classificatory information contained in other information sources.
The drug classes used in these information sources are information-rich, often describing multiple attributes. At the same time, the relatively small number of high-level categories to which we were able to assign nearly all the class descriptors suggests that a tractable reference model can be developed. Despite the importance of many other drug characteristics (e.g., storage and handling procedures for warehouse managers and pharmacists, physical description and smell for poison control workers), a six-category reference model describes nearly all the information found in the three sources we studied. Thus, we believe that a clinically relevant, fine-grained, explicit drug classification scheme can be built and maintained without an overwhelming effort.
Our reviewers had detailed but inconclusive discussions on the distinction between Physiological Effect and Therapeutic Intent. For example, classes like thrombolytics’ and anti-emetics’ can be considered as fitting into either category. That is, members of these classes could be grouped together because they cause an action on a particular body system (breaking up blood clots in the cardiovascular system and reducing vomiting in the autonomic nervous system respectively). Another valid interpretation, however, is that drugs in these classes are grouped together because they treat a patient’s condition of thrombosis or vomiting. NDF-RT’s resolution of these tensions likely will require development of specific use cases.
Other terminologies have adopted different strategies to organize drug data. Although MeSH has adopted the same set of organizing categories as NDF-RT (Mechanism of Action, Physiological Effect, and Therapeutic Use), these categories are not orthogonal, and thus classes such as ‘fibrinolytic agents’ and ‘antiemetic agents’ are listed under both Physiological Effect and Therapeutic Use. This structure neatly sidesteps the difficulty we encountered in determining the boundary between these categories. SNOMED CT, similar to the Medicare Part D classes in our study, generally groups drugs according to a targeted body system and certain structural and functional classes.
A notable gap discovered in NDF-RT’s current reference model involves classifying drugs by their formulation or packaging. Our reviewers agreed on 97 such attributes, even more than Mechanisms of Action, in the NDF drug classes (see Table 1). Of these, the majority involve the intended route of administration, as in the following examples:
Anti-infectives, Vaginal
Oral Hypoglycemic Agents
Antineoplastics, Topical
Although NDF-RT does characterize each drug’s dose form, the formulated route is not captured in the model. Reference hierarchies of formulated or intended routes and regulatory status (e.g., for investigational drugs) would increase NDF-RT’s coverage of the NDF drug categories to more than 90%. Both these enhancements could have obvious uses in clinical decision support applications.
Conclusion
Drug classifications in use today are complex and overlapping. They group drugs along multiple axes or dimensions, and individual classes often include more than one kind of information.
NDF-RT’s reference model explicitly captures three quarters of the information found in the NDF drug classes it seeks to supplement. New reference categories describing drug formulation and regulatory status would improve NDF-RT’s semantic coverage of NDF drug category information to more than 90%.
Disentangling mixed classifications found in real-world information sources may offer benefits to the developers of structured terminologies and ontological resources. Such an exercise can provide a framework for building the terminology’s upper structure and developing a clear understanding of the domain, leading to increasingly understandable, reproducible and useful modeling decisions.
ACKNOWLEDGEMENTS
This work was supported in part by the United States National Library of Medicine Grant (Rosenbloom, 1K22 LM08576-02).
Footnotes
Although NDF allows products to be placed in up to two classes, in practice all products belong to a single class.
References
- 1.DailyMed Web site [database on the Internet] Bethesda (MD): National Library of Medicine (US); 2006. [accessed 2006 Mar 11]. Renese®; [about 6 p.]. Available from: http://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?id=57. [Google Scholar]
- 2.SNOMED International [homepage on the Internet] Chicago, IL: College of American Pathologists; 2006. [accessed 2006 Mar 11]. Available from http://www.snomed.org/ [Google Scholar]
- 3.Medical Subject Headings [homepage on the Internet] Bethesda, MD: National Library of Medicine (US); 2006. [accessed 2006 Mar 11]. Available from: http://www.nlm.nih.gov/mesh/ [Google Scholar]
- 4.MedLinePlus Drug Information [database on the Internet] Bethesda (MD): National Library of Medicine (US); 2006. [accessed 2006 Mar 14]. Available from: http://www.nlm.nih.gov/medlineplus/druginformation.html. [Google Scholar]
- 5.Carter JS, Brown SH, Erlbaum MS, Gregg W, Elkin PL, Speroff T, et al. Initializing the VA medication reference terminology using UMLS Metathesaurus co-occurrences. Proc AMIA Symp. 2002:116–20. [PMC free article] [PubMed] [Google Scholar]
- 6.Spackman KA, Campbell KE, Cote RA. SNOMED RT: a reference terminology for health care. Proc AMIA Symp. 1997:640–4. [PMC free article] [PubMed] [Google Scholar]
- 7.Solomon WD, Wroe CJ, Rector AL, Rogers JE, Fistein JL, Johnson P. A reference terminology for drugs. Proc AMIA Symp. 1999:152–6. [PMC free article] [PubMed] [Google Scholar]
- 8.TDE – Apelon’s Terminology Development Environment [homepage on the Internet] Ridgefield, CT: Apelon, Inc; 2006. [accessed 2006 Mar 11]. Available from: http://www.apelon.com/products/tde.htm. [Google Scholar]
- 9.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 10.Lau LM, Lam SH. Applying the desiderata for controlled medical vocabularies to drug information databases. Proc AMIA Symp. 1999:97–101. [PMC free article] [PubMed] [Google Scholar]
- 11.Center for Medicare and Medicaid Services. Summary of H.R. 1 Medicare Prescription Drug, Improvement, and Modernization Act of 2003 [monograph on the Internet] Rockville, MD: Center for Medicare and Medicaid Services (US); 2006. [accessed 2006 Mar 11]. Available from http://www.cms.hhs.gov/MMAUpdate/downloads/PL108-173summary.pdf. [Google Scholar]
- 12.McCutcheon, Tracey. Medicare prescription drug benefit model guidelines [monograph on the Internet] Rockville, MD: United States Pharmacopeia; 2004. [accessed 2006 Mar 11]. Available from http://www.usp.org/pdf/EN/mmg/finalModelGuidelines2004-12-31.pdf. [Google Scholar]
- 13.Brown SH, Elkin PL, Rosenbloom ST, Husser C, Bauer BA, Lincoln MJ, et al. VA National Drug File Reference Terminology: a cross-institutional content coverage study. Medinfo. 2004;11(Pt. 1):477–81. [PubMed] [Google Scholar]
- 14.Chute CG, Carter JS, Tuttle MS, Haber M, Brown SH. Integrating pharmacokinetics knowledge into a drug ontology: as an extension to support pharmacogenomics. Proc AMIA Symp. 2003:170–4. [PMC free article] [PubMed] [Google Scholar]