Abstract
This paper evaluates a system, UBMedTIRS, for retrieval of medical images. The system uses a combination of image and text features as well as mapping of free text to UMLS concepts. UBMedTIRS combines three publicly available tools: a content-based image retrieval system (GIFT), a text retrieval system (SMART), and a tool for mapping free text to UMLS concepts (MetaMap). The system is evaluated using the ImageCLEFmed 2005 collection that contains approximately 50,000 medical images with associated text descriptions in English, French and German. Our experimental results indicate that the proposed approach yields significant improvements in retrieval performance. Our system performs 156% above the GIFT system and 42% above the text retrieval system.
Introduction
Traditionally, retrieval of images is performed by creating a surrogate description of the contents of the image and using this textual description for search purposes. An alternative approach to retrieving images is content-based image retrieval (CBIR). This approach uses automatically extracted features to build a vector representation that can be used to compute similarity between images. There has been a significant amount of research in CBIR during the last 10 years. Commercial systems such as QBIC from IBM, and Virage offer general capabilities for retrieval of images using automatically extracted features. However, application of CBIR to medical images requires special considerations in terms of feature extraction.
This work explores the combination of CBIR methods with text retrieval as well as automatic mapping of free text to a medical ontology. We present the architecture of a system designed for this purpose and a formal evaluation using a large collection of medical images with annotations in English, French and German.
Content Based Image Retrieval
CBIR uses automatically extracted features such as color, texture and shape to build a vector representation that can be used to compute similarities between images. There is a large and growing amount of research in CBIR which has been accumulated over the last 10 years. The research findings indicate that one of the most promising applications for CBIR is medical imaging. Retrieval of medical images requires specialized methods for manipulating images due to the nature of the images in the applied setting. For example, whereas extraction of color-based features is generally an important aspect for CBIR applications, in medical images this could be misleading since most of the images are in black and white and presence of color might be due to the contrast media used to enhance the image.
According to Müller et al.1 formal evaluation of CBIR systems were rare until a couple of years ago. With the exception of two published works -- ASSERT2 and IRMA3 -- most proposed applications of CBIR for medical images either ignore evaluation issues or limit the discussion of effectiveness to presenting selected queries showing only a few retrieved images.
In the last three years ImageCLEFmed, which is part of the Cross Language Evaluation Forum (CLEF) campaign, has been promoting formal evaluation of CBIR applications for medical images4. The ImageCLEFmed track requires participating teams to use a common collection of diverse medical images and a set of topics that contain sample images as well as text. The systems are required to return a list of images ranked according to similarity to the given set of topics. The ImageCLEFmed task started as a formal track in CLEF 2004 and has attracted a large number of participants from all over the world.
As suggested by Müller et al.1 there are three areas that could benefit from using CBIR methods combined with text retrieval: teaching, research and diagnostics. An immediate application of CBIR in teaching consists of searching interesting cases to present to students or to help students to identify cases with similar images, which would facilitate learning by contrasting the differences and similarities of such cases. CBIR could be used to enhance access to clinical research data. There is also the potential for applying data mining techniques to clinical data in order to expand the knowledge and understanding of various domain sources. Finally, it is possible to envision the use of this type of system in the clinical settings as an application of evidence-based medicine and computer aided diagnosis. For example, the system could help a clinician to find cases that have similar images as the one being diagnosed (similar retrieval), or find difference between the current case and “normal” images (dissimilar retrieval).
Combining Image and Text Features
Traditionally, retrieval of images has been done by searching text associated to them. A common approach that has been employed by librarians is to create a surrogate document that contains metadata associated to the image. This metadata includes a physical description (i.e. format, source of origin, etc) as well as descriptive terms that are usually selected from a controlled vocabulary. This approach has the disadvantage of being labor intensive and might not capture the full extent of the contents of the image. CBIR was proposed as an alternative to get a more exhaustive indexing of the image feature. However, these automatic features cannot capture the semantic content of the image. For this reason we use a combined approach that tries to take advantage of both methods to improve ranking of retrieved images.
The system that we present in this paper was developed by our team for the 2005 ImageCLEFmed track5 and combines three tools that are publicly available. The general guiding principle in our approach is the use of automatic relevance feedback, which has been successfully applied in information retrieval, and the aggregation of evidence from multiple sources. UBMedTIRS (University at Buffalo’s Medical Text and Image Retrieval System) combines the GNU Image Finding Tool (GIFT)6, the SMART retrieval system7, and MetaMap8 in a single interface that allows retrieval of images using multimedia queries that could contain text and/or images.
GIFT is an open source CBIR system developed by the Viper group on Multimedia Information retrieval at the University of Geneva. It allows query by example on images and have retrieval feedback capabilities.
SMART is an information retrieval system developed by Gerard Salton and his collaborators at Cornel University7. It has been used extensively and has proven to perform quite well for text retrieval. The version we used has been modified to include additional weighting schemes (such as pivoted length normalization and Okapi Bm25) and support for 11 European languages.
MetaMap8 is a tool that maps free text to concepts in the Unified Medical Language System (UMLS). The UMLS includes the Metathesaurus, the Specialist Lexicon, and the Semantic Network9. The UMLS Metathesaurus combines more that 79 vocabularies. Among these vocabularies is the Medical Subject Headings (MeSH), and several vocabularies that include translations of medical English terms to 13 different languages.
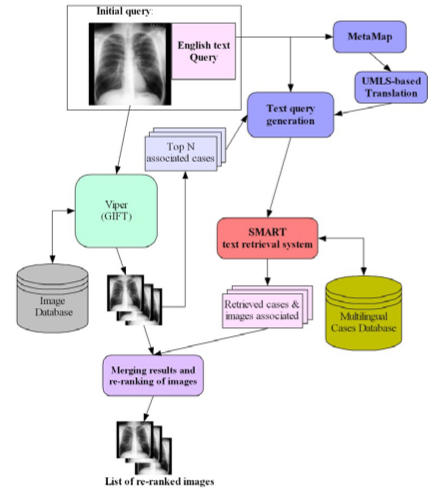
Figure 1 shows the architecture of UBMedTIRS. Queries submitted to the system could consist of an image, text or both. Images are sent to GIFT for processing and matching against the image database. Every image in the database has a text description that can include terms from a controlled vocabulary as well as free text (i.e., dictations generated by physicians or radiologists). The text associated to the top n retrieved images is used to expand the original query. This query expansion step uses Rocchio’s formula to compute the weight of each term as follows:
Figure 1.
Architecture of UBMedTIRS
where α and β are coefficients that control the contribution of the original query terms and the terms from the related image descriptions, respectively. R is the number of text descriptions considered for expansion.
Our system supports multilingual annotations but assumes that the language of the original query is English. The text part of the original query is processed using MetaMap to find the UMLS concepts present in it. These concepts can be used for generating expanded terms as well as for finding translations in French and German. The final text query is built by processing the original query using a simple stemmer, which normalizes plurals, and adding expansion terms generated from the case descriptions of the top retrieved images and from the concept mapping to UMLS.
This query is used by SMART to retrieve the relevant image descriptions. The final step consists in combining the list of images retrieved by the GIFT with the images associated to the case descriptions retrieved by SMART. We use the following formula to compute the final score of each image:
where Ik is the score of the image in the CBIR system and Tk is the score of the image in the text retrieval system. λ and μ are parameters that control the contribution of each system to the final score. Note that this formula assumes that the scores from both systems are normalized between 0 and 1.
Data Collection
We used the ImageCLEFmed 2005 collection which includes approximately 50,000 images and text descriptions4. This collection is a compilation of 4 sources: Casimage, Pathology Educational Institutional Resource (PEIR), Mallinkrodt Institute of Radiology (MIR), and Pathology Images (PathoPic). Table 1 summarizes the details of the image collection. Although the collection includes mostly radiological images (x-rays, CT, MRI, etc), it also includes photographs, power point slides and illustrations. Each image has an associated annotation in at least one of the three languages (English, French and German).
Table 1.
ImageCLEFmed 2005 collection
| Collection | Images | Annotations | Language |
|---|---|---|---|
| Casimage | ~9K | ~2K cases | 80%French
20%English |
| PEIR | ~33K | Per image annotation | English |
| MIR | ~2K | Annotations per case | English |
| PathoPic | ~9k | Annotation per image | German & English |
The ImageCLEFmed collection also includes 25 topics with one to three images and a text describing the information needed. These 25 topics can be organized in three different groups of queries:
Queries that are expected to be solvable using visual features (topics 1–12)
Queries where both text and image features are expected work well (topics 13–23)
Semantic queries where the visual features are not expected to improve results (topics 24 and 25)
Topic relevance judgments were compiled during the ImageCLEF 2005 by pooling the top 100 results of the 12 participating groups and assessing the relevance of each pooled set of image to the corresponding topic.
Collection Preparation and Indexing
We used the SMART system to create a multilingual database with all the case descriptions of the 4 collections of images (Casimage, MIR, Pathopic, and PEIR). We used a common stop list that includes English, French, and German stop words.
Indexing of the images was done using the standard settings of GIFT with 4 grey levels and the full HSV space using the Gabor filter responses in four directions and at three scales.
Experimental Results
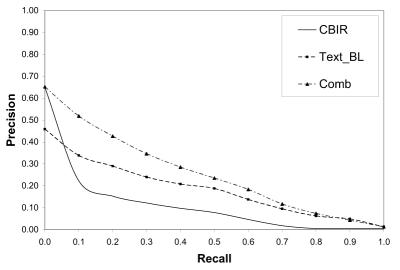
We use two baseline systems for our retrieval experiments. The first baseline corresponds to a run that uses only the image retrieval from GIFT. The second baseline uses only the text from the original topics to retrieve case descriptions and ranking the associated images. Table 2 and Figure 2 show the average precision on all 25 queries at the 11 standard recall points. These runs were generated using the top 10 retrieved documents as relevant and adding the top 50 terms ranked according to Rocchio’s formula using α =8 and β =64. For combining the results we use λ =1 and μ =3. These parameters were derived by using the ImageCLEFmed 2004 data. The results show that the baseline that uses text only performs 80% above the CBIR baseline and that the combination of text and image performs above both baselines (156% above CBIR and 42% above the text baseline). All differences are statistically significant.
Table 2.
Average Precision at Standard Recall Levels
| Recall Levels | CBIR | Text | Comb |
|---|---|---|---|
| 0.0 | 0.6495 | 0.4590 | 0.6519 |
| 0.1 | 0.2222 | 0.3385 | 0.5186 |
| 0.2 | 0.1530 | 0.2902 | 0.4267 |
| 0.3 | 0.1215 | 0.2401 | 0.3467 |
| 0.4 | 0.0976 | 0.2082 | 0.2853 |
| 0.5 | 0.0780 | 0.1879 | 0.2356 |
| 0.6 | 0.0461 | 0.1379 | 0.1836 |
| 0.7 | 0.0183 | 0.0950 | 0.1171 |
| 0.8 | 0.0053 | 0.0625 | 0.0741 |
| 0.9 | 0.0053 | 0.0488 | 0.0430 |
| 1.0
|
0.0053
|
0.0130
|
0.0129
|
| Mean AvgP | 0.0956 | 0.1728 | 0.2451 |
Figure 2.
Recall Precision Graph
Table 3 shows the mean average precision of each system on the three groups of queries described in the data collection. It is interesting to note that the combination of text and images outperforms both baselines in all three groups.
Table 3.
Mean Average Precision on Groups of Queries
| Query group | CBIR | Text | Comb |
|---|---|---|---|
| Visual | 0.1193 | 0.1357 | 0.2352 |
| Visual &Text | 0.0781 | 0.1685 | 0.2238 |
| Semantic | 0.0504 | 0.4192 | 0.4214 |
Analysis and Discussion
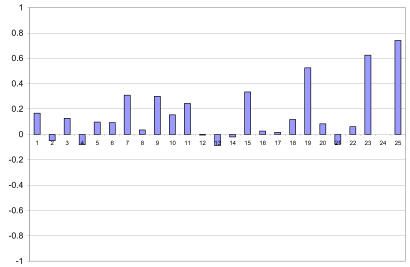
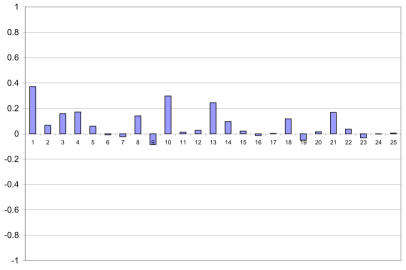
Our experimental results show that the combination of text and image significantly improves mean average precision. In this section we report a detailed query by query analysis. Figure 3 shows the difference in performance of each of the 25 queries with respect to the CBIR baseline. This graph shows more clearly that the combination actually improves the average precision for 16 of the 25 queries. Figure 4 shows the difference with respect to the Text baseline. We can see that the combination improves performance in 14 of the 25 queries. It is interesting to note that most of the gains are obtained in the queries from the visual group (first 12 queries). This confirms that these queries can benefit from using image features.
Figure 3.
Query by query difference in performance with respect to the CBIR baseline
Figure 4.
Difference in performance with respect to the Text baseline
Since this is a task that involves cross-language retrieval we also assessed the effectiveness of our translation method using UMLS to identify English concepts to find the corresponding French and German terms. For this purpose we evaluated the performance of the system using the manual translations provided for each topic. The best mean average precision that we could get for the manual translation queries using text only is 0.1879 and 0.2539 for the combination of text and images. This means that our cross language run achieves 91% of the monolingual performance for text only and 96% of the combined text and image runs. The high values of these percentages are due in part to the fact that most of the documents in the collection are in English (which is our starting language).
Conclusion
We have presented a method that takes advantage of text and image features to improve retrieval of medical images. Our experimental results show that this method yields significant improvements with respect to using text only or image only.
Our current system is able to handle a multilingual collection in English, French and German and the results indicate that the translation method using MetaMap to find English concepts to find the appropriate French and German versions is effective.
We plan to continue exploring the optimization and tuning of the system parameters to different types of queries and also to explore interactive retrieval of medical images in educational and clinical settings.
Acknowledgements
This research was funded in part by an AT&T research grant for junior faculty members of the School of Informatics at UB. We also want to thank the National Library of Medicine for supporting the author of this paper as a visiting researcher during the summer of 2005. Finally I would like to thank Dr. Alan Aronson and the Indexing Initiative team members at NLM for their feedback and for making available the MetaMap program for this research.
References
- 1.Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medicine - clinical benefits and future directions, International Journal of Medical Informatics. 2004;73:1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Shyu CR, Brodley C, Kak A, Kosaka A, Aisen A, Broderick L. ASSERT: A physician-in-the-loop content-based retrieval system for HRCT image databases. Computer Vision and Image Understanding. 1999;75(1/2):111–132. [Google Scholar]
- 3.Keysers D, Dahmen J, Ney H, Wein BB, Lehmann TM. Statistical framework for model-based image retrieval in medical applications. Journal of Enlectronic Imaging. 2003;12(1):59–68. [Google Scholar]
- 4.Clough P, Müller H, Deselaers T, et al. The CLEF 2005 Cross-Language Image Retrieval Track. In: Peters C, Magni R, editors. Cross Language Evaluation Forum 2005. Vienna, Austria: Springer; 2006. [Google Scholar]
- 5.Ruiz ME, Srikanth M. UB at CLEF2004 Cross Language Medical Image Retrieval; Paper presented at: Fifth Workshop of the Cross-Language Evaluation Forum (CLEF 2004), 2005; Bath, England. [Google Scholar]
- 6.GNU Image Finding Tool (GIFT) 2004. http://www.gnu.org/software/gift.
- 7.Salton G, editor. The SMART Retrieval System: Experiments in Automatic Document Processing. Englewood Cliff, NJ: Prentice Hall; 1971. [Google Scholar]
- 8.Aronson A. Effective Mapping of Biomedical Text to the UMLS Metathesaurus: The MetaMap Program. Paper presented at: American Medical Informatics Association Annual Symposium; 2001. [PMC free article] [PubMed] [Google Scholar]
- 9.NLM. 2004. U.S. National Library of Medicine: Unified Medical Language System (UMLS) [PubMed] [Google Scholar]