Abstract
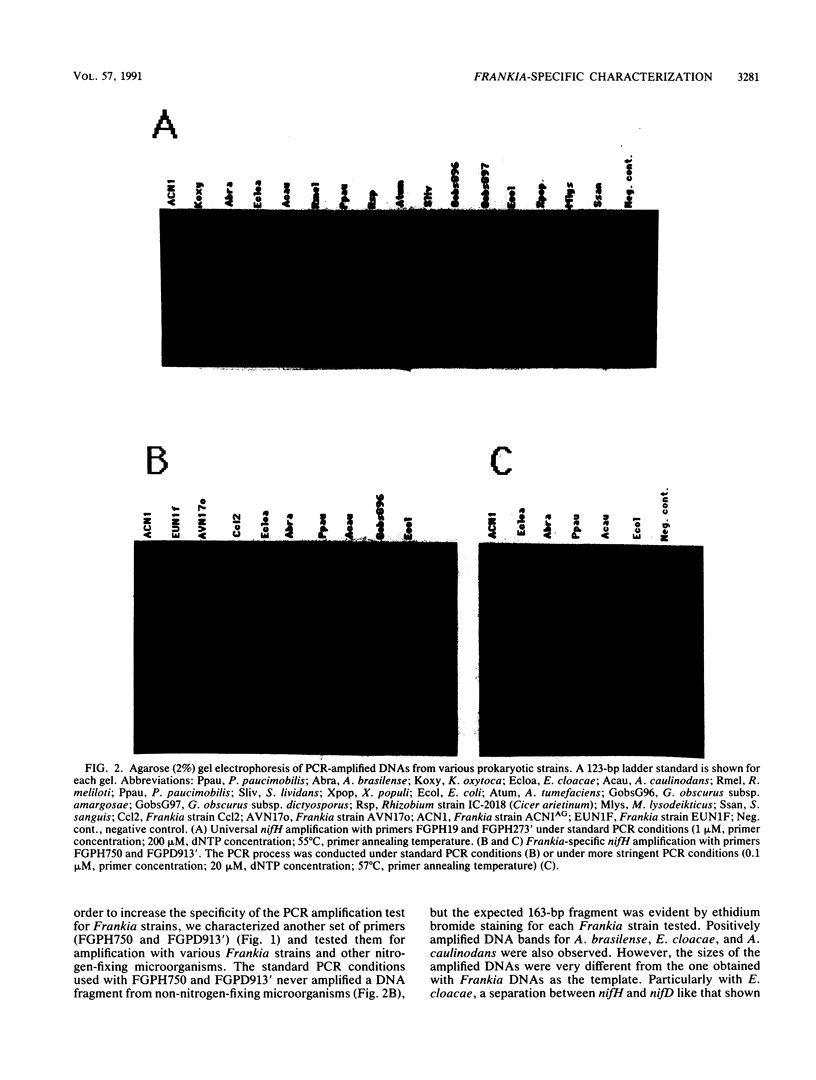
The polymerase chain reaction (PCR) is an in vitro procedure for primer-directed enzymatic amplification of specific template nucleic acid sequences. In order to determine whether a given actinomycete isolated from an actinorhiza (nodule) belongs to the genus Frankia or is a contaminant, we have developed a test based on the PCR. Primers complementary to sequences of two DNA regions corresponding to the nif genes (nifH and nifD) and the rRNA genes (16S and 23S) were specifically chosen to differentially amplify DNAs from Frankia strains but not those from other microorganisms. A series of positive and negative controls were set up by using universal or selective primers resulting in a discriminant amplification, which could be detected after agarose gel electrophoresis. In the nif region, degenerate oligonucleotide primers were used to amplify a target common to all the nitrogen-fixing microorganisms tested, while another set of primers amplified a target with a high specificity for Frankia strains. In the rRNA gene region, universal and specific primers were characterized and tested with DNAs from a wide range of microorganisms. The efficiency of this rapid and sensitive PCR assay was tested with an isolate obtained from Alnus nepalensis nodules, confirming results obtained by nodulation tests.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brenner D. J., McWhorter A. C., Knutson J. K., Steigerwalt A. G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982 Jun;15(6):1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigle K. E., Newton W. E., Dean D. R. Complete nucleotide sequence of the Azotobacter vinelandii nitrogenase structural gene cluster. Gene. 1985;37(1-3):37–44. doi: 10.1016/0378-1119(85)90255-0. [DOI] [PubMed] [Google Scholar]
- Charfreitag O., Stackebrandt E. Inter- and intrageneric relationships of the genus Propionibacterium as determined by 16S rRNA sequences. J Gen Microbiol. 1989 Jul;135(7):2065–2070. doi: 10.1099/00221287-135-7-2065. [DOI] [PubMed] [Google Scholar]
- Embley T. M., Smida J., Stackebrandt E. Reverse transcriptase sequencing of 16S ribosomal RNA from Faenia rectivirgula, Pseudonocardia thermophila and Saccharopolyspora hirsuta, three wall type IV actinomycetes which lack mycolic acids. J Gen Microbiol. 1988 Apr;134(4):961–966. doi: 10.1099/00221287-134-4-961. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M., Hennecke H. Rhizobium japonicum nitrogenase Fe protein gene (nifH). J Bacteriol. 1984 Jun;158(3):1005–1011. doi: 10.1128/jb.158.3.1005-1011.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M., Pirouz T. Numerical classification of sporoactinomycetes containing meso-diaminopimelic acid in the cell wall. J Gen Microbiol. 1982 Mar;128(3):503–527. doi: 10.1099/00221287-128-3-503. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Kaluza K., Fuhrmann M., Hahn M., Regensburger B., Hennecke H. In Rhizobium japonicum the nitrogenase genes nifH and nifDK are separated. J Bacteriol. 1983 Aug;155(2):915–918. doi: 10.1128/jb.155.2.915-918.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanvinde L., Sastry G. R. Agrobacterium tumefaciens Is a Diazotrophic Bacterium. Appl Environ Microbiol. 1990 Jul;56(7):2087–2092. doi: 10.1128/aem.56.7.2087-2092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc D. J., Lee L. N. Physical and genetic analyses of streptococcal plasmid pAM beta 1 and cloning of its replication region. J Bacteriol. 1984 Feb;157(2):445–453. doi: 10.1128/jb.157.2.445-453.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevarech M., Rice D., Haselkorn R. Nucleotide sequence of a cyanobacterial nifH gene coding for nitrogenase reductase. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6476–6480. doi: 10.1073/pnas.77.11.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Nazaret S., Cournoyer B., Normand P., Simonet P. Phylogenetic relationships among Frankia genomic species determined by use of amplified 16S rDNA sequences. J Bacteriol. 1991 Jul;173(13):4072–4078. doi: 10.1128/jb.173.13.4072-4078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., Hendriks L., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P., Bousquet J. Phylogeny of nitrogenase sequences in Frankia and other nitrogen-fixing microorganisms. J Mol Evol. 1989 Nov;29(5):436–447. doi: 10.1007/BF02602914. [DOI] [PubMed] [Google Scholar]
- Normand P., Simonet P., Bardin R. Conservation of nif sequences in Frankia. Mol Gen Genet. 1988 Aug;213(2-3):238–246. doi: 10.1007/BF00339587. [DOI] [PubMed] [Google Scholar]
- Pretorius I. M., Rawlings D. E., O'Neill E. G., Jones W. A., Kirby R., Woods D. R. Nucleotide sequence of the gene encoding the nitrogenase iron protein of Thiobacillus ferrooxidans. J Bacteriol. 1987 Jan;169(1):367–370. doi: 10.1128/jb.169.1.367-370.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinto C., De La Vega H., Flores M., Leemans J., Cevallos M. A., Pardo M. A., Azpiroz R., De Lourdes Girard M., Calva E., Palacios R. Nitrogenase reductase: A functional multigene family in Rhizobium phaseoli. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1170–1174. doi: 10.1073/pnas.82.4.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson R., Woodley P., Jones R. Second gene (nifH*) coding for a nitrogenase iron protein in Azotobacter chroococcum is adjacent to a gene coding for a ferredoxin-like protein. EMBO J. 1986 Jun;5(6):1159–1163. doi: 10.1002/j.1460-2075.1986.tb04341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J. P., Waitches G. M., Scolnik P. A. A DNA fragment hybridizing to a nif probe in Rhodobacter capsulatus is homologous to a 16S rRNA gene. Gene. 1986;48(1):81–92. doi: 10.1016/0378-1119(86)90354-9. [DOI] [PubMed] [Google Scholar]
- Scott K. F., Rolfe B. G., Shine J. Biological nitrogen fixation: primary structure of the Rhizobium trifolii iron protein gene. DNA. 1983;2(2):149–155. doi: 10.1089/dna.1983.2.149. [DOI] [PubMed] [Google Scholar]
- Scott K. F., Rolfe B. G., Shine J. Nitrogenase structural genes are unlinked in the nonlegume symbiont Parasponia rhizobium. DNA. 1983;2(2):141–148. doi: 10.1089/dna.1983.2.141. [DOI] [PubMed] [Google Scholar]
- Simonet P., Le N. T., Du Cros E. T., Bardin R. Identification of frankia strains by direct DNA hybridization of crushed nodules. Appl Environ Microbiol. 1988 Oct;54(10):2500–2503. doi: 10.1128/aem.54.10.2500-2503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet P., Normand P., Moiroud A., Bardin R. Identification of Frankia strains in nodules by hybridization of polymerase chain reaction products with strain-specific oligonucleotide probes. Arch Microbiol. 1990;153(3):235–240. doi: 10.1007/BF00249074. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Charfreitag O. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol. 1990 Jan;136(1):37–43. doi: 10.1099/00221287-136-1-37. [DOI] [PubMed] [Google Scholar]
- Sundaresan V., Ausubel F. M. Nucleotide sequence of the gene coding for the nitrogenase iron protein from Klebsiella pneumoniae. J Biol Chem. 1981 Mar 25;256(6):2808–2812. [PubMed] [Google Scholar]
- Tarrand J. J., Krieg N. R., Döbereiner J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 1978 Aug;24(8):967–980. doi: 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- Török I., Kondorosi A. Nucleotide sequence of the R.meliloti nitrogenase reductase (nifH) gene. Nucleic Acids Res. 1981 Nov 11;9(21):5711–5723. doi: 10.1093/nar/9.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Larebeke N., Engler G., Holsters M., Van den Elsacker S., Zaenen I., Schilperoort R. A., Schell J. Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature. 1974 Nov 8;252(5479):169–170. doi: 10.1038/252169a0. [DOI] [PubMed] [Google Scholar]
- Wang S. Z., Chen J. S., Johnson J. L. The presence of five nifH-like sequences in Clostridium pasteurianum: sequence divergence and transcription properties. Nucleic Acids Res. 1988 Jan 25;16(2):439–454. doi: 10.1093/nar/16.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]