Abstract
Electronic medical records (EMR) constitute a valuable resource of patient specific information and are increasingly used for clinical practice and research. Acronyms present a challenge to retrieving information from the EMR because many acronyms are ambiguous with respect to their full form. In this paper we perform a comparative study of supervised acronym disambiguation in a corpus of clinical notes, using three machine learning algorithms: the naïve Bayes classifier, decision trees and Support Vector Machines (SVMs). Our training features include part-of-speech tags, unigrams and bigrams in the context of the ambiguous acronym. We find that the combination of these feature types results in consistently better accuracy than when they are used individually, regardless of the learning algorithm employed. The accuracy of all three methods when using all features consistently approaches or exceeds 90%, even when the baseline majority classifier is below 50%.
Introduction
Electronic medical records (EMR) constitute a valuable resource of patient specific information and are increasingly used for clinical practice and research. The adoption rates of EMRs across the United States are on the rise and are projected to reach their maximum by the year 2024 (1). In order to maximize the utility of the EMR for care delivery and research, it is important to “unlock” the information contained in the text of clinical reports that are part of the EMR.
Acronyms present a challenge to retrieving information from the EMR. The problem is that many acronyms are ambiguous with respect to their full form or sense. Liu et al. (2) show that 33% of acronyms listed in the UMLS in 2001 are ambiguous. In a later study, Liu et al. (3) demonstrate that 81% of acronyms found in MEDLINE abstracts are ambiguous and have on average 16 senses. In addition to problems with text interpretation, Friedman et al. (4) also point out that acronyms constitute a major source of errors in a system that automatically generates lexicons for medical Natural Language Processing (NLP) applications.
Most previous work on acronym disambiguation has focused on supervised machine learning approaches where a corpus of text is annotated for acronyms and their sense (i.e., expansion) is manually disambiguated. Machine learning algorithms are then trained on this labeled data to generate models, and future ambiguous instances of the acronyms are disambiguated using these models. The disambiguation is typically cast as a classification problem with a known set of senses to be predicted as the class value. This is the approach we adopt in this study, where we view acronym disambiguation as a special case of the word sense disambiguation (WSD) problem. This allows us to rely on features and supervised learning algorithms that are known to work well on that problem (e.g. (5,10,11,12,13,14)).
However, we are aware that fully supervised approaches are usually not scalable due to the time and effort involved in manually creating sufficient amounts of training data. Thus, our future work will focus on semi-supervised and unsupervised techniques that require smaller amounts of training data, as well as developing methods that gather training data automatically. One possible unsupervised approach is to create and cluster context vectors (6). Also, there is a hybrid class of machine learning techniques for WSD that relies on a small set of hand labeled data to bootstrap a larger corpus of training data (7).
Pakhomov et al. (8, 9) developed a method for collecting training data for supervised machine learning approaches to disambiguating acronyms. The method is based on the assumption that the full form of an acronym and the acronym itself tend to occur in similar contexts. The full form found in the text of clinical reports is then used as if it were an acronym to generate training data. Pakhomov et al. (9) have developed a small manually annotated corpus of ninei acronyms and their senses, which was used to test their semi-supervised approach to data generation. They also showed that traditional fully supervised approaches achieve a very high level of accuracy (> 90%).
The objective of this study is three-fold. One is to expand and evaluate the corpus of manually annotated acronyms to include seven additional acronyms for a total of 16. The second is to create a benchmark of accuracy results achieved with three fully supervised machine learning algorithms that can be used as a point of comparison with other approaches to acronym disambiguation. The third is to experiment with several combinations of feature selection methods and learning algorithms in order to determine which are most advantageous for acronym disambiguation.
Materials and Methods
DATASET
We have created a corpus of 7,738 manually disambiguated instances of 16 ambiguous acronyms. This corpus is derived from the Mayo Clinic database of clinical notes. Nine of the acronyms were annotated previous to this study, while the remaining seven were newly annotated for this work. The annotation was carried out the same way in both cases, the only difference being the current process was based on the entire database of 17 million notes spanning years 1994–2005, while the previous one was based on a 1.7 million note subset from 2002.
Table 1 summarizes the data for the 16 acronyms. The acronym and the total number of instances are shown in the first column, where those from the previous study are underlined. Then the top three expansions and associated instance counts (N) are shown in the second and third column. If there are more than three expansions for an acronym, then we combine the counts of the remaining expansions into a single row. We show the percentage of the total instances that have a given expansion in the fourth column.
Table 1.
Distribution of Acronym Expansions
| Acr. | Top 3 Expansions | N | (%) |
|---|---|---|---|
|
AC 464 |
Acromioclavicular
Antitussive with Codeine Acid Controller 10 more expansions |
146
139 109 70 |
31.47
29.96 23.49 15.08 |
|
APC 376 |
Argon Plasma Coagulation
Adenomatous Polyposis Coli Atrial Premature Contraction 10 more expansions |
157
94 55 70 |
41.76
25.00 14.63 18.62 |
| LE
615 |
Limited Exam
Lower Extremity Initials 5 more expansions |
291
270 44 10 |
47.32
43.90 7.15 1.62 |
|
PE 519 |
Pulmonary Embolism
Pressure Equalizing Patient Education 12 more expansions |
251
160 48 60 |
48.36
30.82 9.24 11.56 |
| CP
578 |
Chest Pain
Cerebral Palsy Cerebellopontine 19 more expansions |
321
110 88 59 |
55.54
19.03 15.22 10.21 |
| HD
254 |
Huntington's Disease
Hemodialysis Hospital Day 9 more expansions |
142
75 22 15 |
55.91
29.52 8.66 5.91 |
|
CF 710 |
Cystic Fibrosis
Cold Formula Complement Fixation 6 more expansions |
530
101 36 43 |
74.65
14.22 5.07 6.06 |
| MCI
344 |
Mild Cognitive Impairment
Methylchloroisothiazolinone Microwave Communications, Inc. 5 more expansions |
269
34 18 23 |
78.20
9.88 5.23 6.69 |
| ID
574 |
Infectious Disease
Identification Idaho Identified 4 more expansions |
450
105 7 7 5 |
78.4
18.29 1.21 1.21 0.87 |
|
LA 488 |
Long Acting
Person Left Atrium 5 more expansions |
385
53 17 33 |
78.89
10.86 3.48 6.76 |
| MI
690 |
Myocardial Infarction
Michigan Unknown 2 more expansions |
590
96 2 2 |
85.51
13.91 0.29 0.29 |
|
ACA 541 |
Adenocarcinoma
Anterior Cerebral Artery Anterior Communication Artery 3 more expansions |
473
62 3 3 |
87.43
11.46 0.006 0.006 |
| GE
591 |
Gastroesophageal
General Exam Generose General Electric |
521
40 22 8 |
88.15
6.77 3.72 1.35 |
|
HA 509 |
Headache
Hearing Aid Hydroxyapatite 2 more expansions |
470
30 6 3 |
92.34
5.89 1.18 0.59 |
| FEN
80 |
Fluids, Electrolytes and Nutrition
Drug Fen Phen Unknown |
78
1 1 |
97.50
1.25 1.25 |
|
NSR 405 |
Normal Sinus Rhythm
Nasoseptal Reconstruction |
401
4 |
99.01
0.99 |
METHODS
The objective of our experiments is to compare the efficacy of three different machine learning algorithms: the naïve Bayes classifier, the C4.5 decision tree learner, and a Support Vector Machine, when used with four types of feature sets: unigrams, bigrams, part-of-speech tags, and the combination of all three of these. In our experiments we perform 10-fold cross-validation (using disjoint folds) for each of the 16 acronyms for each of the possible combinations of the three learning algorithms and four feature sets. We also report results for the Majority classifier, which assigns the most frequent expansion to all of the instances of an acronym. This serves as a baseline above which any reasonable method would be expected to perform.
The naïve Bayes classifier has a long history of success in word sense disambiguation (e.g., 10, 11). It is the simplest method we employ, since it assumes the parametric form of a model of conditional independence (that describes the relationships among the features) and so only needs to learn the parameters of this model. Decision trees are a part of this study since they too have a long history in word sense disambiguation (e.g, (5, 13)). They are a generalization of decision lists, which were successfully utilized by Yarowsky (12). Pedersen (13) later explored decision trees with bigram features, and found them to perform at high levels of accuracy. Finally, Support Vector Machines have fared well in recent comparative evaluations of supervised word sense disambiguation systems (e.g., (14)). We use the implementations of these learning algorithms provided by the Weka Data Mining suite (17) and maintain their default configuration settings.
FEATURES
We identify four sets of features for our experiments:
Part-of-Speech (POS) tags
The part-of-speech of the two words to the left and the two words to the right of the acronym to be expanded (the target) are used as features, as is the part-of-speech tag of the acronym itself. We use a modified version of the Brill part-of-speech tagger created by Hepple (16), which provides 55 POS tags including punctuation. This is distributed as part of the ANNIE system in the General Architecture for Text Engineering (GATE) toolkit. Part-of-speech tags are commonly used as features in WSD, and have been found to provide a surprising amount of disambiguation information on their own (e.g., (16)).
Unigrams
Individual words that appear five or more times in the training examples for an acronym are considered as features. These must occur in a flexible window centered around the target acronym that extends five positions to the left and five positions to the right. A flexible window skips low frequency or stop words found near the target acronym, so the flexible window can be understood as the first five high frequency content words that appear to the left and to the right of the target acronym, no matter how far they are from the target. However, this flexible window is confined to a single clinical note, although it does cross sentence and section boundaries if necessary. Stop words are function words (articles, prepositions, etc.) that do not provide meaningful content on their own. We use a manually created list of 107 stop words consisting mainly of propositions, pronouns, auxiliary verbs and proper nouns denoting calendar items.
Bigrams
In our experiments, bigrams are two consecutive words that occur together five or more times. Neither of the words in a bigram may be found in the stop list that we employ for unigrams. A flexible window of five is used for bigrams as well, which means that five significant bigrams to the left and right of the acronym (within the same clinical note) are used as features.
Unigrams + bigrams + POS
The fourth set of features combines all of the above 3 sets – POS tags, unigrams and bigrams into one larger set of features.
Results and Discussion
There were a number of questions that motivated these experiments. First, we were interested to see if the different types of features would result in significantly different performance when used with several different learning algorithms. Second, we were interested to see the effect of the distribution of the senses in the acronyms on our overall results. Third, we wanted to characterize the effect of the flexible window method of selecting lexical features. Finally, we wanted to compare the results of these methods with those reported in (9), at least for the eight acronyms in common between the two studies.
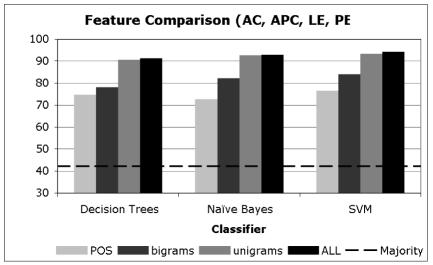
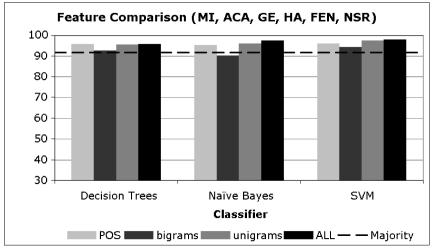
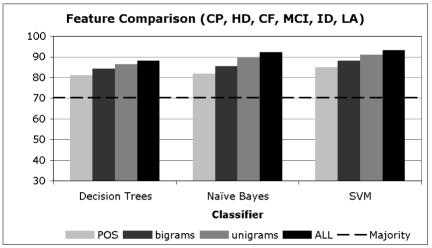
In Charts 1–3, we present averaged results for the different learning algorithms when trained with data consisting of the different types of features we utilized. The acronyms in these charts are selected based on the distribution of the majority sense in the data, where Chart 1 includes the acronyms that have a majority sense of less than 50%, Chart 2 includes those that have a majority sense between 50% and 80%, and Chart 3 includes acronyms with a majority sense over 80%.
Chart 1.
Disambiguation Accuracy of Acronyms with majority sense < 50%
Chart 3.
Disambiguation Accuracy of Acronyms with majority sense > 80%
Chart 2.
Disambiguation Accuracy of Acronyms with majority sense > 50% and < 80%
From Charts 1–3, it is clear that regardless of the distribution of the majority sense, the overall disambiguation accuracy attained is typically at or above 90%. This means that the high level of performance does not depend on having a skewed distribution of senses (with a high majority sense) but rather these methods are carrying out their disambiguation task at very high levels of accuracy regardless of the baseline from which they start.
It is also clear that for all methods the unigram features and the combination of all features provides the highest level of performance. In each of the charts, there is little or no difference between the unigram results and the all-features result, suggesting that the unigram features are providing much of the disambiguation accuracy in the combined feature set.
We do note one interesting difference in the performance of these methods based on their majority sense baseline. For those acronyms with a less pronounced majority sense (Chart 1), note that the POS tags and the bigrams significantly lag behind the unigrams and combined features. This continues to a lesser extent for those acronyms with somewhat more pronounced majority senses (Chart 2), and then disappears for those with very skewed majority senses (Chart 3). We believe that this shows quite clearly that unigrams and the combined feature set are in general superior to the POS tags and bigrams, since they are providing more disambiguation accuracy in the more difficult case where the majority sense is relatively low. To be fair, it is likely that bigrams did not perform well due to the fact that they were selected using a frequency cutoff, when in general more informative bigrams can be selected using measures of association such as Pearson‘s Chi-Squared test or the Log-likelihood Ratio (13). Finally, it is interesting that POS tags have performed at a high level of accuracy on their own, suggesting that a significant percentage of acronym ambiguity can be resolved using syntactic information only.
In Charts 1–3, it is interesting to note that the different learning algorithms perform at very similar levels of accuracy when they are using the same features. While there are differences between them, they are smaller than the differences observed for a single algorithm when using different features. For example, the accuracy when using POS tags with the three algorithms in Chart 1 varies from 71% to 76%, while the difference between POS tags and unigrams for SVMs varies from 76% to 91%. This indicates that the key to attaining high accuracy is not the choice of the learning algorithm, but rather the features.
An intuitive way to see this in Charts 1–3 is to notice that the differences between the bars of the same shade (feature) across the three methods is much less (generally) than the differences observed among the different features within a single method. In fact, these differences are statistically significant. Using a two-tailed paired t-test with α =0.05, the improvement of unigrams over POS tags and bigrams and that of all-features over POS tags, bigrams and unigrams across all the 16 acronyms is significant with p < 0.01.
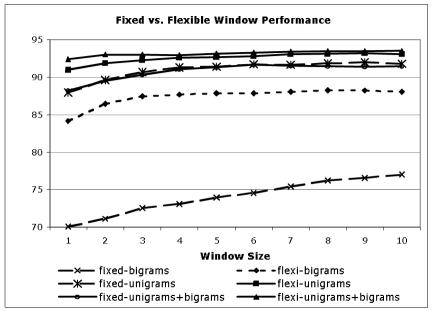
The flexible window that we have introduced in this paper is a novel way of formulating sets of lexical features. We experimented with various different sizes of windows using different feature types, and summarize those results in Chart 4.
Chart 4.
Effect on disambiguation accuracy of increasing the size of the flexible window for feature selection
We see the average accuracy across all three methods for each of the different types of features indicated. We note first that bigrams in a fixed window performed significantly worse than all other measures, and the use of the flexible window significantly improved upon bigrams. For example, a flexible window of size five results in accuracy of 87% for bigrams, while a fixed window of size five achieved an accuracy of 74%. We note smaller but still significant (α =0.05, p < 0.001) improvements for unigrams and a combination of unigrams and bigrams when moving from a fixed window to a flexible one.
We believe that a flexible window as we propose offers significant advantages over fixed windows, in that the number of features that will occur in a fixed window can vary to a large extent depending on syntactic and lexical variations. As such, the flexible window allows us to capture exactly the number of features that we believe to be important. In general we have found that flexible window sizes of three to five tend to work well in practice.
Finally, in Table 2 we compare our results with those of Pakhomov, et al. (9) on the eight acronyms that are common between our studies. The best results obtained by Pakhomov were using a Maximum Entropy model, except for CF which attained its best result with a decision tree.
Table 2.
Comparison of Pakhomov et al. (9) with one of our methods (SVM with all features)
| Acr. | Pakhomov | SVM |
|---|---|---|
| AC | 96.7 | 95.5 |
| ACA | 97.0 | 97.6 |
| APC | 95.9 | 92.3 |
| CF | 95.8 | 97.3 |
| HA | 95.8 | 96.1 |
| LA | 94.6 | 96.7 |
| NSR | 99.0 | 99.0 |
| PE | 93.3 | 92.7 |
In Table 2 we underline and italicize those values where a difference of one percentage point or greater in the methods is observed. In general both sets of results are quite good, and certainly represent acceptable performance.
Conclusions
We have developed a corpus of 16 acronyms manually annotated with their expansions, and established baseline disambiguation performance with three machine learning schemes – the naïve Bayes classifier, decision tree learner and Support Vector Machine. This work significantly extends our previous efforts by creating a validated set of benchmark measurements of the accuracy of fully supervised approaches. This provides a foundation for future work with semi-supervised and unsupervised approaches to acronym disambiguation. We demonstrate a significant improvement in accuracy by using a flexible window for lexical features. We have also further validated our previous findings that a relatively small amount of manually annotated clinical text data can result in very high accuracy of acronym disambiguation. This is encouraging as it may provide a methodology for applications to prospective identification of patients with specific conditions where high accuracy on a limited number of acronyms is required.
Acknowledgements
We would like to thank our annotators Barbara Abbott, Debra Albrecht and Pauline Funk. This work was supported in part by the NLM Training Grant (T15 LM07041-19) and the NIH Roadmap Multidisciplinary Clinical Research Career Development Award (K12/NICHD)-HD49078. Dr. Pedersen has been partially supported in carrying out this research by a National Science Foundation Faculty Early CAREER Development Award (#0092784).
Footnotes
Only eight of these nine acronyms were actually used in the study by Pakhomov et al. (9). The annotation of the ninth acronym (HD) was completed after publication.
References
- 1.Ford E, Menacheni N, Phillips T. Predicting the Adoption of Electronic Health Records by Physicians: When Will Health Care be Paperless? Journal of American Medical Informatics Association. 2006;13(1):106–112. doi: 10.1197/jamia.M1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Lussier Y, Friedman C. American Medical Informatics Association Symposium. Washington, DC: 2001. A Study of Abbreviations in UMLS; pp. 393–397. [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H, Aronson A, Friedman C. American Medical Informatics Association Symposium. San Antonio, TX: 2002. A Study of Abbreviations in MEDLINE Abstracts; pp. 464–8. [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman C, Liu H, Shagina L, Johnson SB, Hripcsak G. American Medical Informatics Association Symposium. Washington, DC: 2001. Evaluating the UMLS as a Source of Lexical Knowledge for Medical Language Processing; pp. 189–93. [PMC free article] [PubMed] [Google Scholar]
- 5.Black E. An Experiment in Computational Discrimination of English Word Senses. IBM Journal of Research and Development. 1988;32(2):185–94. [Google Scholar]
- 6.Purandare A, Pedersen T. Conference on Natural Language Leanring. Boston, MA: 2004. Word Sense Discrimination by Clustering Contexts in Vector and Similarity Spaces; pp. 41–48. [Google Scholar]
- 7.Hearst M. Noun Homograph Disambiguation using Local Context in Large Text Corpora. 7th Annual Conference of the University of Waterloo Center for the new OED and Text Research; Oxford. 1991. pp. 1–22. [Google Scholar]
- 8.Pakhomov S. Semi-Supervised Maximum Entropy Based Approach to Acronym and Abbreviation Normalization in Medical Texts. 40th Meeting of the Association for Computational Linguistics; Philadelphia, PA. 2002. pp. 160–7. [Google Scholar]
- 9.Pakhomov S, Pedersen T, Chute CG. American Medical Informatics Association Annual Symposium. Washington, DC: 2005. Abbreviation and Acronym Disambiguation in Clinical Discourse; pp. 589–93. [PMC free article] [PubMed] [Google Scholar]
- 10.Gale W, Church K, Yarowsky D. A Method for Disambiguating Word Senses in a Large Corpus. Computers and the Humanities. 1992;26:415–39. [Google Scholar]
- 11.Mooney R. Comparative Experiments on Disambiguating Word Senses: An Illustration of the Role of Bias in Machine Learning. Proceedings of the Conference on Empirical Methods in Natural Language Processing; Philadelphia, PA. 1996. pp. 82–91. [Google Scholar]
- 12.Yarowsky D. Unsupervised Word Sense Disambiguation Rivaling Supervised Methods. 33rd Annual Meeting of the Association for Computational Linguistics; Cambridge, MA. 1995. pp. 189–96. [Google Scholar]
- 13.Pedersen T. A Decision Tree of Bigrams is an Accurate Predictor of Word Sense. Second Annual Meeting of the North American Chapter of the Association for Computational Linguistics; Pittsburgh, PA. 2001. pp. 79–86. [Google Scholar]
- 14.Lee YK, Ng HT, Chia TK. Supervised Word Sense Disambiguation with Support Vector Machines and Multiple Knowledge Sources. Senseval-3: Third International Workshop on the Evaluation of Systems for the Semantic Analysis of Text; Barcelona, Spain. 2004. pp. 137–140. [Google Scholar]
- 15.Hepple M. Independence and commitment: Assumptions for rapid training and execution of rule-based. POS taggers 38th Annual Meeting of the Association for Computational Linguistics; Hong Kong. 2000. pp. 278–85. [Google Scholar]
- 16.Mohammad S, Pedersen T. Combining Lexical and Syntactic Features for Supervised Word Sense Disambiguation. Proceedings of the Conference on Computational Natural Language Learning; Boston, MA. 2004. pp. 25–32. [Google Scholar]
- 17.Witten I, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2. San Francisco, CA: Morgan Kaufmann Publishers; 2005. [Google Scholar]