Abstract
The semantic web is envisioned as an evolving set of local ontologies that are gradually linked together into a global knowledge network. Many such local “application” ontologies are being built, but it is difficult to link them together because of incompatibilities and lack of adherence to ontology standards. “Reference” ontologies are an emerging ontology type that attempt to represent deep knowledge of basic science in a principled way that allows them to be re-used in multiple ways, just as the basic sciences are re-used in clinical applications. As such they have the potential to be a foundation for the semantic web if methods can be developed for deriving application ontologies from them. We describe a computational framework for this purpose that is generalized from the database concept of “views”, and describe the research issues that must be solved to implement such a framework. We argue that the development of such a framework is becoming increasingly feasible due to a convergence of advances in several fields.
Introduction
The semantic web is emerging as the most promising long-term solution to the problem of data and computational model integration at the level of meaning. The vision of the semantic web is that local ontologies describing entities and relations relevant to specific application domains will gradually be linked together into world-wide knowledge networks. Recognition of the importance of such ontologies for biomedical data integration has resulted in an increasing number of ontologies of relatively circumscribed scope, which are designed for specific biomedical sub domains. In fact, an important clearing house for these efforts is the Open Biomedical Ontologies (OBO) project [1], which currently houses a growing number of ontologies, from fields such as Zebrafish biology, murine developmental anatomy, and many others.
Virtually all these application ontologies have been or are being developed by domain experts for use in specific types of applications. As such they generally do not conform to principles that permit them to be easily linked to other ontologies in the evolving semantic web [2]. As more ontologies are developed, this problem of incompatible ontologies is becoming reminiscent of the very data integration problem that ontologies are intended to solve.
In this paper we propose an approach to one aspect of this problem, which is to use reference ontologies as a basis for deriving application ontologies. We also propose a computational framework for embedding these derived ontologies in an evolving semantic web, and discuss some of the research problems that must be solved in order to realize such a framework.
Reference ontologies
The idea of a reference ontology, rooted in our own efforts to develop the Foundational of Anatomy (FMA) [3] (http://fma.biostr.washington.edu), is now gaining acceptance by the biomedical informatics community as an ontology type that is distinct from the application ontologies currently in use [4]. Unlike application ontologies, reference ontologies are not designed for any specific application, but are intended to be re-used in multiple application contexts. To-date there is only one biomedical reference ontology in existence, the FMA, but others are in development through the OBO Foundry Project [5]. Ideally, each of these reference ontologies will encompass one of the fields of basic medical science, and just as basic science knowledge is re-used in multiple ways in research and clinical practice, so too will reference ontologies be re-used by including segments of one or more of them in different application ontologies. Since reference ontologies are a relatively new development they are being designed as extensions or specializations of high-level ontologies that take a global view of multiple domains of reality, and do so in accordance with principles of ontology science [4, 6]. Therefore, application ontologies that are based on reference ontologies should be more easily linked together in the semantic web than application ontologies developed de novo.
However, the promise of reference ontologies will only be realized if ways can be found to utilize them in specific applications. Because they are meant to be reused, reference ontologies are broad and deep, whereas application ontologies are narrow and shallow. Reference ontologies are designed according to strict ontological principles [2–4, 6], whereas application ontologies are designed according to the viewpoint of an end-user in a particular domain. The result of these differences is that reference ontologies are too large and detailed to be used “out-of-the box” in applications, even when developers are aware of them and would like to use them.
These issues lead to the following two specific research problems that must be addressed in order to realize the potential of reference ontologies as a foundation for the semantic web. The first problem is how to generate application ontologies from one or more reference ontologies. Rather than developing ad hoc application ontologies, we would like to develop formal methods for specifying the transformation from reference ontologies to application ontologies. The methods should be specified in a declarative way (rather than as ad hoc programs) so that they may easily be re-run as the source ontologies change, and so that the specification may be generated and manipulated by graphical interfaces.
The second problem is how to provide access to these application ontologies through query interfaces rather than as downloadable files. This problem must be solved in order to link large ontologies into the semantic web, but on a more immediate timescale it arises because of the issue of version control: the reference ontology changes after someone has built an application ontology from it. A solution to this problem is to never actually deliver the application ontology to application developers, but instead to make it available as a web service that can be queried by web-enabled applications [7]. Such an approach should greatly reduce the versioning problem, since the query interface will always have access to the most up-to-date version of the reference ontology.
The view-based approach
Our approach to these problems is based on the concept of views that is prevalent in the database world. In database terminology, a view is a query that computes a new table from old tables. In our framework we extend this notion to ontologies, in which an application ontology becomes a view of a reference ontology (or another application ontology that is itself a view of reference ontology). The view is defined as a query expressed in a formal ontology query language (like SQL in the relational database world). The advantages of this approach are 1) the view definition (application ontology) is specified via a set of queries, and hence can be manipulated by another program such as a graphical interface; and 2) the application ontology is always up-to-date since it is non-materialized (or virtual) – that is, it is only defined by the set of queries constituting the view. At any time the view can be materialized by running the queries and saving the file, which can then be made available in the format needed by the application. However, an increasing number of applications are designed with service oriented architectures (SOAs). In these cases materializing the view is unnecessary, since the applications can access the view via web services. In this manner the “virtual” application ontology is never out of date because the queries always directly access the reference ontology from which it is derived.
Computational framework
In order to implement the view-based approach we have designed the computational framework shown in its simplest form in Figure 1 Figure 1. In this form the system includes 1) A reference ontology R, 2) A wrapper W that dynamically converts R from its internal representation into the representation expected by the query language, 3) a view definition V that defines a non-materialized application ontology A as one or more queries over the wrapped reference ontology, 4) a View Generator application VGen that allows a user to graphically specify the view V, and 5) a View Query Processor VQP that accepts a query Q expressed by an application over the application ontology A, reformulates Q as a query Q’ over R, and returns the results. Both VQP and W are implemented as web services.
Figure 1.
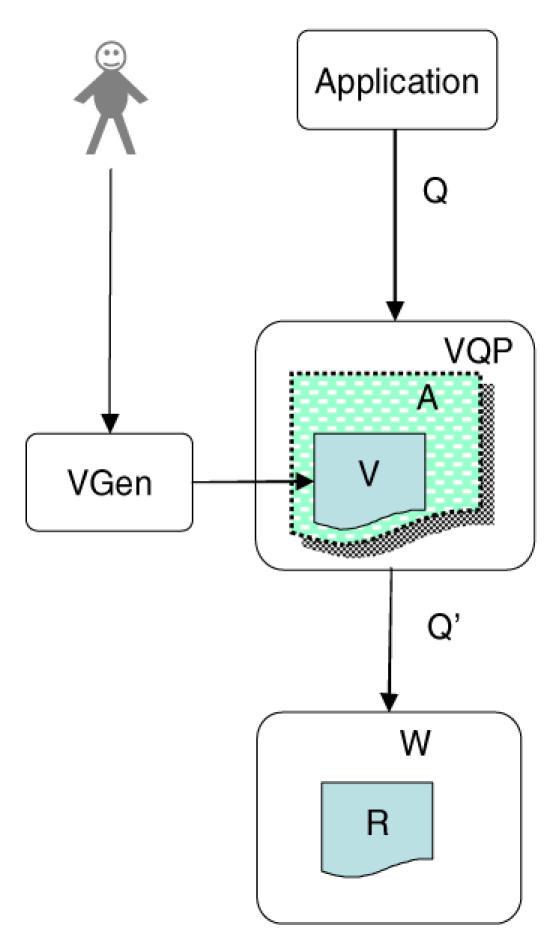
Single source reference ontology
The basic concept is that the combination of a view definition V and an instance of the View Query Processor VQP constitutes virtual application ontology A, that is, a non-materialized view of the underlying reference ontology.
As an example, R might be the FMA, a reference ontology that includes over 75,000 nodes and 2 million relations mirroring the structure of the entire body. However, a neuroscientist may like to see an application ontology A that only includes that portion of the FMA that deals with the parts of the brain, and that vastly simplifies the set of relationships between these neuroanatomical concepts. Thus, in this case the view V needs to select only those portions of the FMA that are brain structures, and then needs to add, remove or rename existing links between these structures in order to generate a simplified partonomy.
A query Q over this neuroanatomy application ontology A might ask for the parts of the temporal lobe of the brain. The instance of VQP associated with A would compose Q with the query V expressing the transformation from R to A, in order to generate a new query Q’, which would be sent to the wrapper W. The wrapper would in turn convert Q’ to a query over the underlying representation of R, which in the case of the FMA is SQL, since the FMA is stored in a relational database for efficiency.
A more complex implementation of the framework is shown in Figure 2. This case shows that both VQP and the wrappers accept the same query language over the same type of virtual ontology representation, and that a view V can define a distributed query over multiple source ontologies. Continuing the previous example, the neuroanatomy application ontology from Figure 1 Figure 1 is shown at the bottom of Figure 2 within the dotted rectangle as application ontology A1 derived from reference ontology R1, the FMA.
Figure 2.
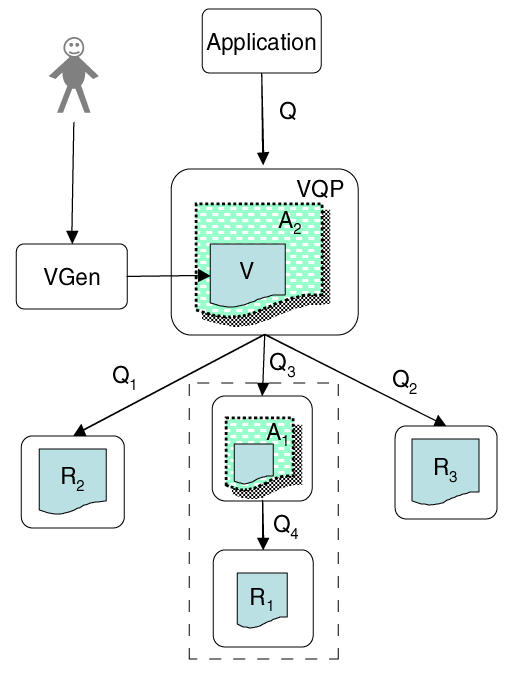
Multiple source ontologies
A new application ontology A2 might be derived by combining elements from A1 (neuroanatomy) a second reference ontology R2 of radiology imaging modalities (MRI, CT, PET), and a third reference ontology R3 of pathological processes (cancer, inflammation). In accordance with the desiderata for reference ontologies [4] we assume that R1, R2 and R3 are disjoint, and therefore in general cannot be mapped to each other.
A2 might represent an ontology of pathological brain anatomy, as seen in various imaging modalities, for use by an image annotation program for clinical images. In this case the VGen program would need to allow the user to generate the view V by grabbing and combining elements of all three of the source ontologies A1, R2 and R3. Query Q issued by the image annotation application might be something like, find all brain tumor types that arise in any part of the temporal lobe and that are visible by MRI. The association between tumor, location and imaging modality would be defined by the view V. Thus, VQP would need to compose Q with V in order to reformulate Q as a series of queries Q1, Q2, and Q3 over each of the separate source ontologies R2, R3 and A1. The query Q3 over A1 would in turn need to be reformulated as query Q4 over R1. The joined results from these queries would be returned to the application, which might then display them as a list of tumor types, organized by location and imaging modality, which could be selected from in order to annotate a specific clinical image.
Figure 3 shows that the ability of VQP to access more than one source ontology, each of which may either be a wrapped reference ontology or another instance of a non-materialized application ontology, gives this framework a fractal property that allows arbitrarily complex webs of interacting, non-materialized ontologies to be gradually built up. In addition, since any application ontology may be materialized and wrapped in a wrapper, and since existing materialized application ontologies may also be accessed through wrappers, the framework allows for the kind of gradual interlinking of ontologies that is a salient part of the vision of the semantic web.
Figure 3.
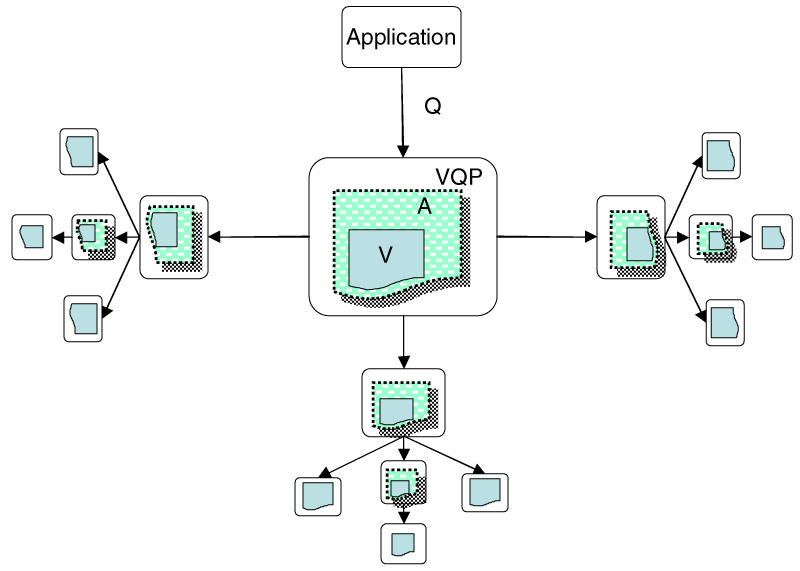
Fractal nature of the framework
Research issues and related work
The key research issues implied by this framework are: 1) how to specify the view V needed to derive an application ontology from possibly more than one source ontology, 2) how to build a web service (VQP) that accepts queries over the application ontology and reformulates them as queries over the underling reference ontology/ies, 3) how to make the process of answering queries efficient enough to be useful, and 4) how to design a graphical interface (VGen) so biologists can specify the views without having to learn the complex view definition language (VDL) that will be needed.
Since we are designing this system to be part of the semantic web we assume that reference ontology R is either directly represented in a semantic web language such as RDF/S or OWL or can be converted by wrapper W to appear as RDF/S/OWL. Since there are as yet very few OWL query languages, and since every OWL representation is a valid RDF representation, we initially assume that the ontologies are either represented in RDF/S or can be wrapped to appear as RDF/S. Given these choices the research issues become specific to the semantic web:
1. Specifying the view V
The first issue is the selection of an RDF/S query language. Several such languages have been developed [8], the most relevant for our purposes being SparQL and RQL. Although SparQL is close to becoming a web standard it does not yet permit complex regular expressions over link paths (eg., find all parts of parts of the brain transitively to a predetermined level of granularity). RQL, on the other hand, can be more easily extended to handle such regular expressions.
The second issue is the choice of a view definition language (VDL) expressed in the underlying RDF query language. A large amount of work has been done in the database community to develop SQL or XQuery-based view definition languages [9, 10], in contrast to a relatively modest effort in the ontology community [11–13]. One reason for the possible choice of RQL as an RDF query language is the existence of RVL, a View Definition Language built in RQL [14].
Given the choice of query and view definition language (VDL), the next task is to extend VDL to handle the complex transformations that define the mapping between source and target ontologies. The extensions will need to include a query component for efficiently selecting those RDF triples (source node, target node and link) that need to be included in the target ontology, and a view definition component that specifies how the triples need to be transformed.
2. Building the View Query Processor
The problem is illustrated in Figure 1. Given query Q over application ontology A, VQP will need to compose Q with the complex query V defining the view over reference ontology R, computing a new query Q’ = Q o V that can be processed by the wrapper W. If R is expressed in another representation than RDF/S, then W will in turn need to reformulate Q’ into a series of queries over the internal representation of R, which for the FMA is a relational database.
The problem is further complicated in Figure 2
Figure 2, in which query Q must be reformulated as a series of distributed queries over multiple source ontologies, some of which may in turn need to further reformulate the queries, ending up eventually at materialized reference or application ontologies. Query reformulation has been extensively studied in the database community, both within relational databases [15], and for converting between XQuery and SQL [10, 16]. Thus, many of the techniques from these areas should be applicable to query reformulation over RDF/S/OWL ontologies.
3. Achieving efficiency
It is highly likely that long chains of query reformulations, if not optimized for efficiency, will generate response times that are too slow for interactive use. Again, the database community has extensive experience in query optimization that could prove useful for ontologies [10]. Example techniques include schema optimization, removal of common sub expressions, view composition, and caching of materialized views at various points along the reformulation chain.
4. Facilitating the specification of views by users
Our own experience with XQuery convinces us that most biologists will not want to learn a complex View Definition Language (VDL) in order to specify the mappings between source and target ontologies. Thus, the View Generator (VGen) will need to implement a graphical user interface that allows a user to select one or more source ontologies, visualize each of them as a graph, select subsets of each source, and define the transformations of these subsets into the target. The GUI should allow the effects of these mappings to be immediately visible in the target, and should include methods for zooming in on large ontologies that cannot be visualized all at once. Although a significant amount of work has been done in visualizing small ontologies, much less work has been done in visualizing larger ontologies [17].
Discussion
This paper advocates for the use of reference ontologies as a foundation for the semantic web, and proposes a computational framework for realizing such a foundation. Although significant research issues must be solved before the framework can become a reality, such a reality seems increasingly possible due to a convergence of advances in ontology science, ontology representation and query languages, Internet bandwidth, web services and service oriented architecture, database research in view definition and query reformulation, and the establishment of a national center for ontology research.
Acknowledgements
Funded by NIH grants DC02310 and LM007714
References
- 1.OBO. Open Biomedical Ontologies. 2005. http://obo.sourceforge.net.
- 2.Zhang S, Bodenreider O. Law and order: Assessing and enforcing compliance with ontological modeling principles in the Foundational Model of Anatomy. Computers in Biology and Medicine. 2005;36(7–8):674–693. doi: 10.1016/j.compbiomed.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosse C, Mejino JLV. A reference ontology for bioinformatics: the Foundational Model of Anatomy. Journal of Bioinformatics. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. http://sigpubs.biostr.washington.edu/archive/00000135/ [DOI] [PubMed] [Google Scholar]
- 4.Burgun A. Desiderata for domain reference ontologies in biomedicine. J Biomed Inform. 2006;39(3):307–313. doi: 10.1016/j.jbi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Open Biomedical Ontologies. OBO Foundry. 2006. http://obofoundry.org/
- 6.Rosse C, Kumar A, Mejino JLV, Cook DL, Detwiler LT, Smith B. Proceedings, AMIA Fall Symposium. Washington, D.C: 2005. A strategy for improving and integrating biomedical ontologies; pp. 639–643. http://sigpubs.biostr.washington.edu/archive/00000176/ [PMC free article] [PubMed] [Google Scholar]
- 7.Dameron O, Noy NF, Knublauch H, Musen MA. Accessing and manipulating ontologies using web services. The Semantic Web - IWSC 2004: Third International Semantic Web Conference. 2004. http://smi-web.stanford.edu/people/noy/papers/dameron2004iswc.pdf.
- 8.Haase P, Broekstra J, Eberhart A, Volz R. A comparison of RDF query languages. 2004. http://www.aifb.uni-karlsruhe.de/WBS/pha/rdf-query/rdfquery.pdf.
- 9.Levy AY, Mendelzon AO, Sagiv Y, Srivastava D. PODS. 1995. Answering queries using views; pp. 95–104. [Google Scholar]
- 10.Fernandez M, Kadiyska Y, Morishima A, Suciu D, Tan W. Silkroute: a framework for publishing relational data in XML. ACM Transactions on Database Technology. 2002;27(4) [Google Scholar]
- 11.Miklos Z, Neumann G, Zdun U, Sintek M. Querying semantic web resources using TRIPLE views; Second International Semantic Web Conference; Sanibel Island, Florida. 2003. [Google Scholar]
- 12.Volz R, Oberle D, Studer R. Implementing views for light-weight web ontologies. IEEE Database Engineering and Application Symposium (IDEAS); Hong Kong, China. 2003. [Google Scholar]
- 13.Noy NF, Musen MA. Specifying ontology views by traversal. The Semantic Web - IWSC 2004: Third International Semantic Web Conference; 2004. p. 713. [Google Scholar]
- 14.Magkanaraki A, Tannen V, Christophides V, Plexousakis D. Viewing the semantic web through RVL lenses. Second International Semantic Web Conference; Sanibel Island, Florida: Springer-Verlag. 2003. pp. 96–112. [Google Scholar]
- 15.Halevy AY, Ives ZG, Madhavan J, Mork P, Suciu D, Tatarinov I. The Piazza peer data management system. IEEE Transactions on Knowledge and Data Engineering. 2004;16:787–798. [Google Scholar]
- 16.Bales N, Brinkley J, Lee ES, Mathur S, Re C, Suciu D. A framework for XML-based integration of data, visualization and analysis in a biomedical domain. Proceedings, Third International XML Database Symposium (XSym 2005); Trondheim, Norway. 2005. pp. 207–221. http://sigpubs.biostr.washington.edu/archive/00000178/ [Google Scholar]
- 17.Storey MA, Musen MA, Silva J, Best C, Ernst N, Fergerson RW, Noy NF. Jambalaya: Interactive visualization to enhance ontology authoring and knowledge acquisition in Protege. Workshop on Interactive Tools for Knowledge Capture, K-CAP-2001; Victoria, Canada. 2001. http://www.cs.uvic.ca/~mstorey/papers/kcap2001.pdf. [Google Scholar]


