Abstract
Automated surveillance tools can provide significant advantages to infection control practitioners. When stored in a relational database, the data collected can also be used to support numerous research and quality improvement opportunities. A previously described electronic infection control surveillance system was remodeled to provide multi-hospital support, an XML based rule set, and interoperability with an enterprise terminology server. This paper describes the new architecture being used at hospitals across BJC HealthCare.
Introduction
Manual review of microbiology cultures is a time-consuming process and can leave little time for other responsibilities. Automated surveillance tools have been shown to improve sensitivity over more traditional techniques and reduce the amount of time needed to track infections.1 The infection control data collected for surveillance can also be stored in a clinical data warehouse and used for numerous research and quality initiatives. 2
Barnes Jewish Hospital in St. Louis has been using the GermWatcher™ electronic microbiology surveillance application since 1993.3 Using fixed length text reports, the existing C++ application parsed data from a single facility and classified organisms by applying two sets of hard-coded rules. Data were presented to the users via a client application developed using PowerBuilder™. While effective at one hospital, the original GermWatcher™ design was difficult to migrate to new facilities.
Errors were common when parsing the text reports due to frequent changes in the lab systems. These errors would have been compounded in a multi-hospital deployment. Applying facility specific rules presented another challenge because the rules were hard-coded in the application.
The existing application also lacked the ability to standardize codes from multiple facilities. This was an important consideration to ensure that rules written for one hospital could be applied seamlessly to cultures from other hospitals. Finally, the existing user interface could not provide facility specific content and would have been a challenge to support across multiple hospitals.
Increased interest in the application from other hospitals at BJC HealthCare led to a redesign in 2004 with an enterprise view in mind. The goal was to develop an application that would be flexible enough to meet the needs of all the hospitals in the network.
To accomplish this, we focused on improving several key aspects of the application. First, we replaced the text reports with a generic eXtensible Markup Language (XML) report that could be processed with less risk of failure. We also moved the rules out of the application and into an XML based rule set that could be configured for the needs of an individual facility. By interfacing the new application with the enterprise terminology server, we were able to map individual facility codes to a standard set of enterprise codes. A web-based user interface minimized support issues.
Methods
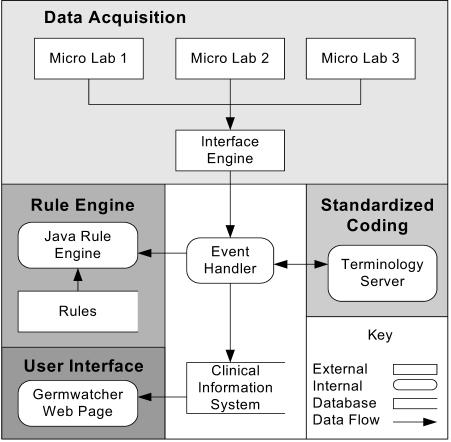
Figure 1 shows the general architecture of the GermWatcher™ application. The key aspects of this architecture will be presented individually.
Figure 1.
General Architecture
Data Acquisition
The first step in re-engineering GermWatcher™ was developing a process to extract microbiology data from different laboratory systems across the enterprise. The original application parsed a fixed length text report and was difficult to maintain. Small changes in the report format caused the application to fail or store faulty data.
Initially we considered using Health Level 7 (HL7) messages, but found that several laboratory systems at BJC could not support HL7 without purchasing additional components from the vendor. Other lab systems that could generate HL7 imbedded descriptive text reports in the messages rather than discrete data.
To leverage work done for the original application, we used each laboratory system’s report writing capabilities to produce a generic XML message that could be parsed more reliably. The XML message schema is more streamlined than HL7 and contains only the information relevant to GermWatcher™. Figure 2 shows the list of fields and attributes available within the body of the microbiology XML messages.
Figure 2.
Microbiology XML Data Elements
XML messages generated by the laboratory software are captured by an interface engine and placed in a queue (IBM MQSeries®) for processing. Messages are generated based on a schedule, and each hospital can define their own schedule according the workflow of the microbiology lab and the needs of the infection control staff. The GermWatcher™ event handler continually monitors the queue, so real-time or near real-time data acquisition is possible. If an error occurs in the parsing process, the event handler automatically routes the invalid message to a separate error queue monitored by the support team.
Standardized Coding
Parsed data from the XML microbiology messages contain facility specific codes for antibiotic susceptibilities, specimen types, organisms, and other common concepts. The Medical Entity Dictionary (MED) maps these facility codes to a standard set of enterprise codes used in the rules for the rules engine. The MED is a terminology server responsible for encoding data for the enterprise.
Antibiotic and specimen strings are mapped to concepts from the Systemized Nomenclature of Human and Veterinary Medicine (SNOMED). Organisms are encoded using concepts from the Unified Medical Language System’s (UMLS) Metathesaurus. Organism strings from each facility are mapped to a Concept Unique Identifier (CUI) when possible, allowing us to take advantage of the relationship information stored in the Metathesaurus to develop more powerful query tools in future phases.
After the data elements are parsed and coded, the event handler packages them for delivery to the rules engine. A separate object is created for each organism in a culture, and these objects are passed to the rule engine one at a time for processing.
Rule Engine
The GermWatcher™ rule engine was developed within BJC HealthCare using the Java Specification Request 000094 (JSR-000094), or Java™ Rule Engine Application Program Interface (API) specification. This provides the framework for assembling a ruleset, passing an object to the rule engine, executing the rules, and returning an object to the calling application.
In our implementation, each rule can be assigned to one or more rulesets, and each facility can have one or more active rulesets in place. A layer called the ‘rule type’ logically groups similar rules together to aid maintenance, but rules are still added or removed from a ruleset individually.
Ruleset information is stored in a relational database, and a single table defines which rules are active at each hospital. Additional dictionary tables provide descriptions of the rules, rule types, and rulesets. Any Open Database Connectivity (ODBC) compliant tool can be used to query and maintain the rulesets.
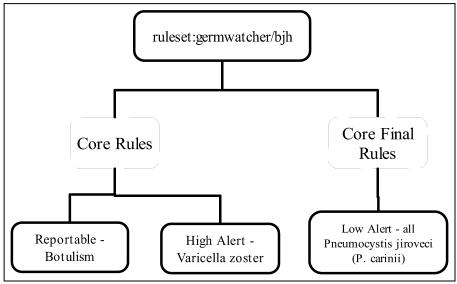
Figure 3 shows how three rules are grouped under a sample ruleset. Both the botulism and Varicella zoster rules are grouped under the “Core Rules” rule type and apply to all cultures regardless of status within the lab. The rule for Pneumocystis jiroveci (P. carinii) is assigned to the “Core Final Rules” rule type and applies only to cultures that are given a final date by the microbiology lab.
Figure 3.
GermWatcher™ Ruleset
JSR-000094 specifications defined the rule engine interface, but did not define the syntax of the rules themselves. After reviewing several options, we based our final design on the reaction rule model described by Boley and colleagues.3 In this model, the rule body contains one or more conditions that are validated before an action is taken. Each single premise or condition is defined as an atom, and multiple atoms can be evaluated using Boolean operators. We consulted the Rule Markup Initiative website for information on syntax and sample rules.5
Figure 4 contains the XML syntax for a Botulism rule. The rule performs a single operation and checks to make sure the organism field matches the enterprise code for C. botulinum. If the check is successful, it updates the classification in the input object to reportable. If the organism field does not match the criteria, rule execution is terminated and the next rule in the sequence is processed.
Figure 4.
Syntax for Botulism Rule
After the rule engine is finished, it returns the input object along with a list of triggered rules. GermWatcher™ stores the triggered rule list and classification results in a Clinical Information System, along with the demographic, culture, and organism data from the XML message.
Each organism can trigger multiple rules, and the organism classification is determined by the rule with the highest priority classification. The organism with the highest priority classification determines the culture classification. Table 1 defines the seven major classifications that can apply to both cultures and organisms. Only the most recent version of a culture is stored to minimize duplicate data.
Table 1.
Classification Definitions
| Classification | Definition |
|---|---|
| Reportable | Preliminary or final lab result; Meets state criteria for reportable disease |
| High Alert | Preliminary or final lab result; Critical for daily workflow |
| Low Alert | Final lab result; Important for ongoing surveillance; Not required for daily workflow |
| Watch | Final lab result; Common skin contaminant; Will be monitored for confirming cultures |
| Pending | Preliminary lab result; No rules triggered |
| Not Significant | Final lab result; Not of interest |
| Negative | Preliminary or final lab result; Negative result |
Following the initial classification and storage of a culture, the rule engine is called again to process a smaller set of temporal rules. These are time-based rules that monitor common skin contaminants (CSCs) such as coagulase-negative Staphylococci for confirming cultures across a registration. For example, one temporal rule confirms a “watched” organism from a blood culture if the same organism is found in another blood culture within a twenty-four hour period.
User Interface
A web-based user interface was developed using Java™ Server Faces and the Jakarta Project’s open source Apache Tomcat. This approach reduces the need for on-site support to install client side applications. A Lightweight Directory Access Protocol (LDAP) server provides user authentication, and patient data are secure behind our firewall.
The user interface sorts the cultures according to the classification and displays only new cultures that have not been reviewed by the infection control staff. Organism classifications are identified by color, which allows easy identification in cultures with multiple organism results. A custom query screen provides infection control practitioners (ICPs) with a way to generate ad-hoc reports from a number of different search criteria, including nursing unit, collection date, organism name, antibiotic susceptibility, and whether an organism is thought to be nosocomial or community acquired.
A secondary application identifies the organisms classified as reportable and populates an electronic version of the Missouri state reportable form. ICPs can manually enter additional information on the electronic form before printing a hard copy to fax to the appropriate destination. Currently we have been unable to send the reportable disease information electronically, but GermWatcher™ could support it once the specifications are agreed upon by state and local agencies.
Results
The new GermWatcher™ architecture was rolled out as a production application in December 2004. It was initially deployed at a large academic hospital in St. Louis, MO and was installed at two community hospitals in the second half of 2005.
As of March 1, 2006, more than 85,000 cultures with over 106,000 discrete organisms have been screened using live data. Each culture requires sub-second processing times, and approximately 1,500 messages pass through the queue each day. This includes both new messages and those messages that are the result of updates to an existing culture
About 110,000 historical cultures with 150,000 discrete organisms were loaded using data from the previous GermWatcher™ application. In total, nearly 250,000 rules have been triggered; Table 2 shows how the organisms were classified according to the highest priority rule. The academic hospital in Table 2 is the largest facility in BJC HealthCare with approximately 900 staffed beds. Community A and Community B have 450 and 350 beds respectively. The academic hospital does not currently receive negative cultures. All organisms at the academic hospital that were classified as negative came as part of a positive culture with at least one positive organism. Development is underway to capture negative cultures for all hospitals running GermWatcher™.
Table 2.
Organism Classification by Hospital
| Academic (Feb 2002) | Community A (Sep 2005) | Community B (Nov 2005) | |
|---|---|---|---|
| Reportable | 2175 | 44 | 73 |
| High Alert | 44021 | 1535 | 992 |
| Low Alert | 79667 | 5036 | 1682 |
| Watch | 14709 | 720 | 284 |
| Pending | 806 | 150 | 93 |
| Not Significant | 68157 | 5332 | 2413 |
| Negative | 4348 | 15115 | 12737 |
| Total | 213883 | 27932 | 18274 |
Discussion
The initial development and release of the new enterprise GermWatcher™ application was a success, and it outperforms the previous version in terms of processing power and maintainability. The new architecture is more than six times faster and will support the increased volume as we roll GermWatcher™ out to more hospitals.
The XML messages have reduced the number of parsing errors, and the error queue makes it easier to identify problems when they are encountered. In the old system, we often had to wait for the infection control practitioners to identify data problems caused by changes in the laboratory report. Now errors are immediately isolated for quick resolution.
From the user perspective, the single biggest gain over the prior manual paper-based system has been the ability to write ad-hoc queries. Similar reports were possible in the absence of GermWatcher™, but relied on the availability of a report writer in each hospital’s microbiology laboratory. There was often a lengthy turn around time for these new reports, which affected the performance of the infection control staff.
The new architecture will also support multi-hospital queries for operational and research agendas. Using the standard enterprise codes, a single query can gather results from several hospitals if the query writer has the correct permissions. ICPs only have access to results from their own facilities through the user interface, but specially trained users will be able to mine data across all hospitals at the request of the corporate infection control department.
During the initial development we encountered a few problems with the design of the rule engine and the XML rules. These were not serious, but required a certain amount of rework to overcome.
The rule engine runs in the same Java™ Virtual Machine (JVM) as the parsing application and receives a Java™ object as input. Our goal with the rule engine was to keep it generic, with no first hand knowledge of the contents of the input object. This made it difficult to perform logical operations on the variables within the Java™ object when the rule engine did not know the data types of the constants defined in the XML rules. To overcome this problem we added a ‘class’ attribute to the XML rule that provides the data type of a constant to the rule engine. The same attribute also provides the object name for XML tags that reference a variable. This allows for more complex, nested input objects if needed.
We also found that some operations were too complex to code using generic relational and Boolean operators. Others operations required database calls to find information not contained within the input object. A <method> tag was added to the rule schema that identifies a custom method defined within the input object. This allows any implementation specific operation to be defined in the input object and passed into the rule engine. The disadvantage of this approach is that we departed from the RuleML model we started with and are instead using a rule schema that is unique to our rule engine.
As we started expanding GermWatcher™ to the second hospital, we encountered three additional challenges that added to the complexity of the project. First, facilities that implemented the same laboratory system could not share the same extraction process because each hospital has flexibility in coding and storing the data within the system. It can take several weeks to customize the extraction for a new facility and requires close cooperation with the technical laboratory team.
Second, the primary users of GermWatcher™ at the initial academic hospital are focused on detecting and preventing nosocomial infections. A number of outpatient tests, including serology based reportable diseases, are not performed in the microbiology laboratory and were never part of the original GermWatcher™ interface. These out-of-scope test results are critical to the workflow of the community hospitals, and we are currently developing an interface to capture and translate out-bound HL7 messages from the general laboratory system to GermWatcher™ compatible XML messages. Tests that are out-sourced to reference laboratories have not been addressed.
Finally, we started the rollout with no formalized plan for resolving differences among facilities. As we installed the application at additional hospitals, we received requests to modify the rules and the user interface to accommodate differences in workflow. Some of the requests were necessary, but others were more superficial. Worse, some of the requests were conflicting and could not be done without making hospital specific user interfaces. The solution was to organize a task force comprised of representatives from each hospital to help set policy and resolve disagreements.
Conclusion
We successfully converted GermWatcher™ from a single facility application to an enterprise application. Users have responded well to the new architecture, and our task force has been effective in organizing and prioritizing the enhancement requests from the infection control practitioners. They are also working to identify a mandatory set of rules for all BJC hospitals. Despite the unforeseen problems encountered during the development and roll out phases, the project has been moving forward and seven additional hospitals are scheduled to implement GermWatcher™ by the end of 2007.
REFERENCES
- 1.Wright MO, Perencevich EN, Novak C, et al. Preliminary assessment of an automated surveillance system for infection control. Infect Control Hosp Epidemiol. 2004 Apr;25(4):325–32. doi: 10.1086/502400. [DOI] [PubMed] [Google Scholar]
- 2.Wisniewski M, Kieszkowski P, Zagorski B, et al. Development of a clinical data warehouse for hospital infection control. J Am Med Inform Assoc. 2003;10:454–62. doi: 10.1197/jamia.M1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn MG, Steib SA, Fraser VJ, Dunagan WC. An expert system for culture-based infection control surveillance. Proceedings of the 17th Annual Symposium on Computer Applications in Medical Care; 1993 Oct 31-Nov 3; Washington, DC. pp. 171–5. [PMC free article] [PubMed] [Google Scholar]
- 4.Boley H, Tabet S, Wagner G. Proceedings of the Semantic Web Working Symposium. Stanford, CA: 2001. Jul–Aug. Design rational of RuleML: A markup language for Semantic Web rules. [Google Scholar]
- 5.The Rule Markup Initiative [homepage on the internet] [[modified 2006 Jan 31; cited 2006 Mar 10]]. Available from: http://www.ruleml.org/