Abstract
WHO-ART was developed by the WHO collaborating centre for international drug monitoring in order to code adverse drug reactions. We assume that computation of semantic distance between WHO-ART terms may be an efficient way to group related medical conditions in the WHO database in order to improve signal detection. Our objective was to develop a method for clustering WHO-ART terms according to some proximity of their meanings. Our material comprises 758 WHO-ART terms. A formal definition was acquired for each term as a list of elementary concepts belonging to SNOMED international axes and characterized by modifier terms in some cases. Clustering was implemented as a terminology service on a J2EE server. Two different unsupervised machine learning algorithms (KMeans, Pvclust) clustered WHO-ART terms according to a semantic distance operator previously described. Pvclust grouped 51% of WHO-ART terms. K-Means grouped 100% of WHO-ART terms but 25% clusters were heterogeneous with k = 180 clusters and 6% clusters were heterogeneous with k = 32 clusters. Clustering algorithms associated to semantic distance could suggest potential groupings of WHO-ART terms that need validation according to the user’s requirements.
Keywords: Unsupervised machine learning, Semantic distance, Pharmacovigilance
INTRODUCTION
The World Health Organization (WHO) collaborating centre for drug safety monitoring collects case reports of suspected adverse drug reactions and stores them in a database, containing over 3.5 million case reports in 2006. A data mining algorithm was developed in order to extract new knowledge on adverse drug reactions (ADR) from this database [1]. This algorithm uses the information component (IC) to measure the strength of the association between a particular drug and a specific ADR. However, the algorithm does not take into account semantics of ADRs coded with the WHO-ART (World Health Organization – Adverse Reaction Terminology) dictionary. The algorithm has a high sensitivity (85%) and a low specificity (44%) [2]
The WHO-ART dictionary describes 32 system organ classes (SOC) [3]. Each class is a hierarchy of terms. The structure is rigid and impedes grouping terms in a flexible way. For instance, other clinical points of view like morphological aspects cannot be handled with the WHO-ART ADR classification system. We assume that clustering of WHO-ART terms describing related medical conditions may help grouping similar case reports before applying data mining algorithms.
We present in this paper a method for clustering semantically related WHO-ART terms thanks to a semantic distance operator that was developed in our laboratory in a different context [4].
BACKGROUND
In a hierarchical structure, semantic distance expresses the similarity between two nodes by counting the minimum number of edges connecting the nodes. Different approaches are proposed to compute this similarity. Some methods include external information like statistics on use of concepts (nodes) in a particular context [5]. Other methods take into account the distance between the node and the root in the structure.
Besides this, unsupervised machine learning algorithms aim at grouping objects into homogeneous classes or clusters. There are two major approaches:
In the hierarchical agglomeration approach, bottom-up algorithms group the closest objects iteratively until there is only one class and top-down algorithms cut sets of objects into two classes iteratively until cutting is no more possible. The general result of these algorithms is a tree structure and clusters are obtained by cutting the tree structure at a specified level.
In the iterative reallocation approach, algorithms start from an initial classification called "centres". Each element is attached to the nearest centre and repeatedly the algorithms replace the centres by the centre of each class obtained previously and each point is associated to the closest new centre. The loop stops when the maximum number of iterations (a priori fixed) is reached or when the centre is stable between two successive iterations. These techniques require specifying the number of clusters.
The major problems in the use of these clustering algorithms are the choice of the most appropriate level to cut the tree structure in the hierarchical agglomeration approach and the choice of the number of clusters for the iterative reallocation. Some researchers proposed a hybrid approach: iterative reallocation is used to provide a finite number of clusters and hierarchical agglomeration is applied to each cluster. A Measure of the probability of an element to belong to a specific cluster is also proposed to improve hierarchical agglomeration [6,7]. This algorithm called pvclust is based on the hclust algorithm for the agglomeration of the data and provides two different measures of the probability.
MATERIAL AND METHODS
Material
We provide formal definitions (FDs) for 758 WHO-ART terms using two knowledge acquisition techniques described in a previous work: 321 terms defined from UMLS, 320 terms defined using morphosemantic analysis and 117 terms defined after expert evaluation [8]. FDs were expressed in the SNOMED International nomenclature [9] which has 11 axes organized in a hierarchical structure. FD of a term is the list of medical conditions attached to this term along the different axes. In some cases, modifier concepts were attached to an element of the FD to give some precision. For example, the WHO-ART concept Acute Pancreatitis is formally defined as Pancreas, NOS (T-65000) + inflammation (M-41000) + Acute (G-A231). In this FD, pancreas is the localisation and belongs to the topographical axis of SNOMED, inflammation is the morphological aspect of the ADR and acute is the modifier concept used to refine the type of inflammation.
Semantic distance
The semantic distance method used in the terminological system is an implementation of the Leacock, Chodorow algorithm [10]: semantic distance of two concepts is the minimum number of edges connecting the two concepts in the semantic network associated to an index of depth of the concepts in the hierarchical structure. In a multi-axial terminological system like SNOMED International, the semantic distance of two concepts is provided by the LP-norm which is the combination of the weight associated to each axis and the semantic distance of every elements of the FD on different axes as shown in the formula below:
| (1) |
A and B are the two terms to be compared, n the number of axes to be considered, Wi the weight of axis i, PAi and PBi projections of A and B on axis i and p a positive number. We choose p = 2 for the Euclidean distance. For example distance between [hepatitis / cholestatic hepatits] is 0.03; [myalgia / arthralgia] is 0.39; [pancreatitis / oesophagitis] is 0.83; [hepatic failure / lactic acidosis] is 1.63.
WHO-ART terms clustering
We use R as statistical environment for unsupervised machine learning [11]. The advantage of such environment is that there is no need to redevelop the implementation of algorithms. First we use the pvclust algorithm, a hierarchical agglomeration algorithm. Second, we use kmeans, an iterative reallocation algorithm. We choose to create 32 and 180 clusters which are respectively the number of system organ classes and the number of high level terms in WHO-ART terminology. The kmeans algorithm is based on the algorithm of Hartigan [12]. The pvclust algorithm provides the approximate unbiased (AU) p-values and bootstrapping probability (BP) values to measure the uncertainty in hierarchical clustering [6,7]. Rserve is used as proxy between the R environment and the application server. Rserve is a TCP/IP server which allows other programs to use facilities of R from various languages without the need to initialize R or link to the R library [13]. It receives data input and parameters for the clustering algorithms and binds the results returned by R environment.
The weight attributed to each axis during semantic distance computation influence the clustering algorithm results. Results obtained with the same weight on each axis are submitted to expert evaluation.
Terminology server
Semantic distance and clustering algorithms are implemented as terminology services on a J2EE (Java 2 Enterprise Edition) platform. This terminology server provides an access to different semantic distance services through web pages: semantic distance computation of two WHO-ART terms, computation of the k nearest neighbours of a term according to the semantic distance, creation of semantic distance matrix file or similarity table.
The terminology server allows choosing the weight of each axis of SNOMED International and the SOCs of interest. The results of semantic distance computation between terms are stored in a file and passed to the R statistical environment for unsupervised machine learning or clustering purposes.
For semantic distance computation and clustering purposes, we use the terminology server. Several functionalities are added: similarity table file creation and binding mechanism for the statistical environment. Similarity table files are used to store semantic distance results and considered as data input for the clustering tasks. Similarity tables contain NxN matrix where N is the total number of terms from the selected SOCs of interest. The weight attributed to each axis of SNOMED International influences the semantic distance value. Weight values depend on the axis which we want to privilege in the clustering. If we want to have concepts clustering according to the SNOMED International topography axis, it is judicious to attribute a high value to this axis. The semantic distance measure does not replace distance measures used in machine learning algorithms but allows converting textual data input into numerical values which can be exploited for machine learning.
RESULTS
Terminology server
The terminology server implementation is based on the JBossTM application server and a MySQLTM database is used for storage. Enterprise Java Beans 2.1 are server side distributed objects written using the Java language. Terminology services are implemented as stateless session beans such as distance(x,y) or similarity table. Entity Beans are persistent objects used to store formal definitions in the MySQLTM database. These are implemented as container managed entity beans and benefit from caching techniques that speed up access to persistent objects. The HTML pages are dynamically generated using Servlet and Java Server Pages technologies. For time consuming tasks like similarity table creation and clustering, tasks are executed in the background and the user is alerted by email when the task is finished.
Hierarchical clustering (pvclust)
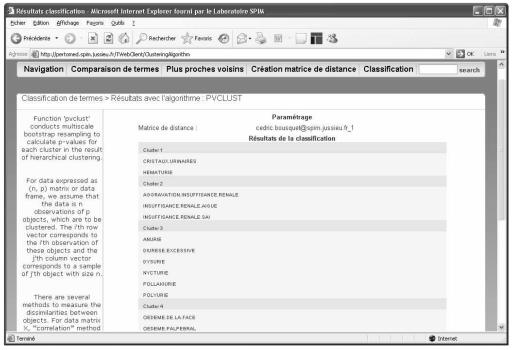
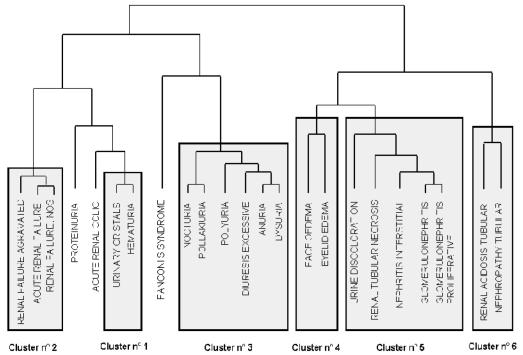
With the "complete" agglomerative method associated to BP = 0.99, 385 terms (50.8%) are grouped in 122 clusters. An example of clustering result with pvclust algorithm is shown on figure 1 (HTML page generated by the server) and figure 2 (dendrogram generated by the server). The query is about urinary system disorders. Six clusters are generated by the algorithm. For example Cluster 1 is {Urinary crystals, Haematuria} and Cluster 2 is {Renal failure aggravated, Renal failure acute, Renal failure NOS}. The evaluation highlights 8 types of clusters:
Figure 1.
HTML page describing clustering of 20 urinary system disorder terms with the pvclust algorithm
Figure 2.
A dendrogram of the clusters is generated by the server
synonym terms (example: {Gingival hypertrophy, Hyperplasia gingival});
antonym terms (example: {Metabolic acidosis, Metabolic Alkalosis});
physiological function abnormality or exacerbation (example: {Dyspnoea, Hyperpnoea });
frequently associated symptoms (example: {Vomiting, Nausea});
abnormal laboratory tests (example: {SGOT increased, SGPT increased});
pathology and its associated aetiology (example:{Haematuria, Urinary crystal});
terms having a close anatomical localization (example: {Pharyngitis, Irritation of throat});
terms expressing a degree of severity of an observed disorder (example: {Coma, State semi comatose}).
Iterative reallocation clustering (kmeans 32)
The kmeans algorithm groups all submitted terms. The number of term per cluster varies from 6 to 92. In many cases, clusters are made of terms coming from different system organ classes of WHO-ART. Experts annotate:
8 heterogeneous (25%) clusters (example: Akinesia, Hypertension, Cough);
4 clusters on anatomical localization (digestive tract, skin…);
8 clusters on pathology (skin pathologies inflammation, ocular pathologies, …)
12 clusters of terms indicating close or same symptoms (pain, fever…).
Iterative reallocation clustering (kmeans 180)
For k= 180 clusters the number of term per cluster varies from 1 to 22. Over the 180 clusters obtained with kmeans algorithm, 12 clusters (6,7%) have no interest for example {Coma - anaesthesia difficult}. The remainders of the clusters are useful. Experts annotate 6 types of clusters:
related to anatomical localization,
related to pathology,
related to symptoms,
terms having the same anatomical localization and pathology,
related to abnormal laboratory tests,
clusters associating the same pathology and clinical signs.
DISCUSSION
We detailed a methodology for clustering WHO-ART formally defined terms. The semantic distance allows applying machine learning algorithms and obtaining clusters of similar WHO-ART terms. The developed terminology server provides an efficient tool to parameterize the different algorithms, to visualize the obtained clusters and to compare the different results provided by the different methods. Using kmeans, 8/32 (25%) clusters were heterogeneous with k = 32 and 12/180 (6.7%) clusters were heterogeneous with k = 180. Distance to the centre of the cluster and number of terms in each cluster were usually smaller using k = 180 than k = 32. Clusters were more homogeneous when choosing k = 180. The hierarchical algorithm was not very useful because half WHO-ART terms were not clustered using this algorithm.
The data model contains elementary concepts enriched or specified by modifiers in some cases. Our semantic distance is smaller when two terms belong to the same SNOMED axis. For example signs and symptoms are usually associated because they belong to the function axis and present no projections on other SNOMED axes. Some clusters were built according to close positions on a single SNOMED axis (for example the topographic or morphologic axes) or according to their projections on two different axes (for example both topographic and morphologic axes). Only a small part of one or two SNOMED axes was involved in each cluster according to their relative projections on each axis.
Perspectives
This work allows exploitation of machine learning algorithms based on similarity of medical concepts expressed in natural language. We plan to investigate what parameters influence the creation of irrelevant clusters: These parameters are related to (1) accuracy and consistency of formal definitions and (2) computation of semantic distance.
Relationship between the elementary concepts is not expressed using explicit relations. A new version of SNOMED terminological system (SNOMED CT) is available and contains additional terms, a richer conceptual model and formal definitions based on description logic that may be useful for improving our FDs. Terms having unsatisfactory FDs were usually assigned to the wrong cluster. Clustering with semantic distance might help identifying errors in FDs that were not detected by the expert evaluation.
Number of clusters with kmeans algorithm where chosen arbitrary according to the current structure of WHO-ART and should be optimized.
This work is part of the EI-Xplore project that aims at improving signal detection in pharmacovigilance. The evaluation of impacts of the clusters on signal detection will be the subject of a later study. Web services functionality will be added in order to link the terminology server to signal detection applications.
Semantic interoperability among distributed and heterogeneous applications means that the communicating parts have a common understanding of the meaning of the data they exchange. Such terminology server may help to match the representation of entities from different controlled vocabularies on the basis of semantic distance [14]. For example we could match equivalent terms from WHO-ART and ICD-10 using our server [8]. We plan to use the server API to develop new applications for data mining and semantic queries in databases.
Acknowledgments
AKNOWLEDGMENTS
We acknowledge the French Agence Nationale de la Recherche (ACI Masses de données) for supporting the EI-Xplore project.
REFERENCES
- 1.Bate A, Lindquist M, Edwards IR, Olsson S, Orre R, Lansner A, De Freitas RM. A Bayesian Neural Network Method for Adverse Drug Reaction signal generation. Eur J Clin Pharmacol. 1998;54(4):315–21. doi: 10.1007/s002280050466. [DOI] [PubMed] [Google Scholar]
- 2.Lindquist M, Stahl M, Bate A, Edwards IR, Meyboom RH. A retrospective evaluation of a data mining approach to aid finding new adverse drug reaction signals in the WHO international database. Drug Saf. 2000;23(6):533–42. doi: 10.2165/00002018-200023060-00004. [DOI] [PubMed] [Google Scholar]
- 3.Uppsala Monitoring Center. The WHO-ART Adverse Reaction Terminology. 2003. [last accessed Jan. 5, 2005]. http://www.umc-products.com/graphics/3149.pdf.
- 4.Bousquet C, Jaulent MC, Chatellier G, Degoulet P. Using semantic distance for efficient coding of medical concept. Proc AMIA Symp. 2000:96–100. [PMC free article] [PubMed] [Google Scholar]
- 5.Resnik P. Using information content to evaluate semantic similarity in a taxonomy. Proceedings of the 14th International Joint Conference on Artificial Intelligence; Montreal. 1995. [Google Scholar]
- 6.Shimodaira H. Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Annals of Statistics. 2004;32:261–4. [Google Scholar]
- 7.Suzuki R, Shimodaira H. An application of multiscale bootstrap resampling to hierarchical clustering of microarray data: How accurate are these clusters?. The Fifteenth International Conference on Genome Informatics; 2004. p. 34. [Google Scholar]
- 8.Iavindrasana J, Bousquet C, Jaulent MC. MIE’2006. Maastricht NL: Knowledge acquisition for computation of semantic distance between WHO-ART terms. Wednesday, 30 August 2006. [PubMed] [Google Scholar]
- 9.Lussier YA, Rothwell DJ, Cote RA. The SNOMED model: a knowledge source for the controlled terminology of the computerized patient record. Methods Inf Med. 1998;37(2):161–4. [PubMed] [Google Scholar]
- 10.Leacock C, Chodorow M. Felbaum C. WordNet: An Electronic Lexical Database. Cambridge, MA: The MIT Press; 1998. Combining local context and WordNet similarity for word sense identification; pp. 265–83. [Google Scholar]
- 11.The R project for Statistical Computing. [last accessed Mar. 10, 2006]. http://www.r-project.org/
- 12.Hartigan J. A. and Wong, M. A. A k-means clustering algorithm. Applied Statistics. 1979;28:100–8. [Google Scholar]
- 13.Rserve - A fast way to provide R functionality to applications. In: Hornik K, Leisch F, Zeileis A, editors. Proc. of the 3rd International Workshop on Distributed Statistical Computing; 2003. [Google Scholar]
- 14.Degoulet P, Sauquet D, Jaulent MC, Zapletal E, Lavril M. Rationale and design for a semantic mediator in health information systems. Meth Inform Med. 1998;37(4–5):518–26. [PubMed] [Google Scholar]