Abstract
Developing computer-interpretable clinical practice guidelines (CPGs) to provide decision support for guideline-based care is an extremely labor-intensive task. In the EON/ATHENA and SAGE projects, we formulated substantial portions of CPGs as computable statements that express declarative relationships between patient conditions and possible interventions. We developed query and expression languages that allow a decision-support system (DSS) to evaluate these statements in specific patient situations. A DSS can use these guideline statements in multiple ways, including: (1) as inputs for determining preferred alternatives in decision-making, and (2) as a way to provide targeted commentaries in the clinical information system. The use of these declarative statements significantly reduces the modeling expertise and effort required to create and maintain computer-interpretable knowledge bases for decision-support purpose. We discuss possible implications for sharing of such knowledge bases.
INTRODUCTION
In recent years, professional societies, health maintenance organizations, and government agencies have produced a flood of clinical practice guidelines (CPGs) for the purpose of disseminating evidence-based best practices to healthcare providers. To deliver context-sensitive or patient-specific guideline content to clinicians at the point of care, a number of groups have developed representation formats—from document-oriented schemas to formal knowledge models—for marking up or encoding CPGs in computer-interpretable formats. Most guideline-modeling formalisms that provide patient-specific decision-support use either rule-based representation or models where the recommendations of a guideline are represented in flowchart-like algorithms that link decisions and actions.1 The processes for selecting, disambiguating, and formalizing recommendations from narrative text to these computer-interpretable rules and algorithms are extremely labor-intensive and require close collaboration between content experts and knowledge engineers. Thus, the resource requirement for developing these computable guidelines remains a serious bottleneck for their wide-spread adoption. Strategies to reduce the necessary effort include the use of general knowledge-editing tools (e.g., Protégé2) and specialized guideline editors (e.g., the GUIDE workbench3). Others have proposed methodologies that require step-wise refinement of guidelines from narrative text to marked-up documents before converting them to computable knowledge bases.4
Instead of encoding guideline recommendations as a large collection of rules or very detailed algorithms, in this paper, we present an alternative approach. We formulate a significant subset of the guideline content in a declarative representation format (1) making statements about concepts and relationships with no flow-of-control or behavioral assumptions, (2) stating relatively simple relationships between patient conditions and possible interventions, and (3) that can be authored and maintained by clinician informaticians with minimal training in the modeling tool. These statements can be used as inputs in more abstract and maintainable rules or algorithms for determining the preferred alternatives in guideline-directed decision-making. Furthermore, these statements can be used independently of guidelines to generate targeted alerting or explanation messages in components of an electronic medical record such as order sets.
This use of declarative statements was developed and validated in two separate efforts to create decision-support systems for guideline-based care. The first, ATHENA DSS (Assessment and Treatment of Hypertension: Evidence-Based Automation Decision Support System), developed and deployed at several Department of Veteran Affairs medical centers, is a system for the treatment of hypertension.5 It uses the EON guideline model,6 developed at Stanford Medical Informatics, Stanford University, as the basis for encoding hypertension guideline knowledge. The second is the SAGE (Standards-Based Sharable Active Guideline Environment) project, a consortium consisting of research groups at GE Healthcare, the University of Nebraska Medical Center, Apelon, Inc., Stanford Medical Informatics, and the Mayo Clinic. SAGE seeks to create the technology for integrating guideline-based decision support into enterprise clinical information systems.
METHOD
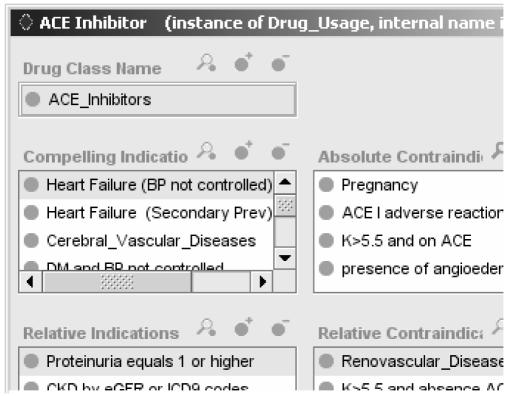
In the EON/ATHENA representation, a key part of the guideline encoding is the enumeration of relationships between patient conditions and drug classes used to manage hypertension. The Sixth Report of the Joint National Committee on Prevention, Diagnosis and Management of Hypertension (JNC 6), the available national guideline when the ATHENA knowledge base was initially developed, categorized considerations for individualizing antihypertensive therapy in terms of relationships such as compelling indication and may have favorable effects on comorbid conditions. The EON representation allows for eight relationships* that are expressed as properties of drug classes. Figure 1 shows a partial screenshot of such an enumeration for ACE Inhibitor as seen in the Protégé knowledge-engineering environment. Each property value (e.g., Heart Failure in the Compelling Indication property) represents a declarative statement linking the drug class with the patient condition through a relationship. Patient conditions are either mapped to codes of the International Classification of Diseases, 9th Revision, through terminological subsumption relationships or are defined using expressions that are evaluated in terms of available data (e.g., Elevated Uric Acid is defined as expressions that check the value of the Uric Acid test result).
Figure 1.
Partial view of the EON/ATHENA representation of relationships (e.g., compelling indication) between patient conditions and drug classes.
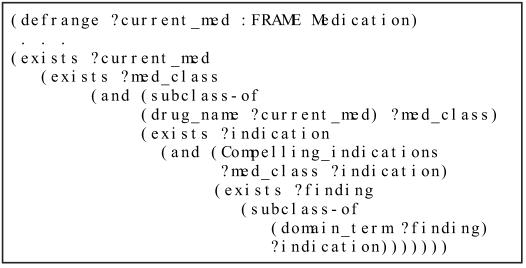
To use these declarative statements in generating guideline-directed therapy recommendations, EON provides a logic-based language, described elsewhere,7 for writing decision criteria that relate patient information to these declarative statements. Figure 2 shows an ATHENA criterion, written in that language, that checks the existence of compellingly indicated drugs among a patient’s medication records.
Figure 2.
ATHENA criterion checking the existence of compellingly indicated medication. The syntax uses a prefix notation. The variable ?current_med ranges over instances of a patient’s medication records, ?med_class ranges over drug classes in the knowledge base, ?indication ranges over possible medical conditions in the knowledge base, and ?finding ranges over instances of patient findings.
The SAGE project generalized the EON/ATHENA representation for declarative statements (1) by defining the concept of Evidence Statements that represent relationships between clinical conditions and interventions along with additional contextual information and supporting references, and (2) by implementing these statements in standardized vocabularies such as SNOMED-CT. Thus, an Evidence Statement encodes a statement such as "In the context of the management of hypertension, presence of heart failure is a compelling indication for the use of ACE inhibitor, with strength of evidence ‘based on randomized controlled trial’ according to the Seventh Report of the Joint National Committee on Prevention, Diagnosis and Management of Hypertension (JNC 7)” (Figure 3). Instead of using a fixed number of properties to relate drug classes and patient conditions, the SAGE representation made relationships such as Compelling Indication terminological codes that can be values of the relationship property of Evidence Statements. Patient conditions such as presence of heart failure are encoded as Boolean expressions that represent patient states.
Figure 3.
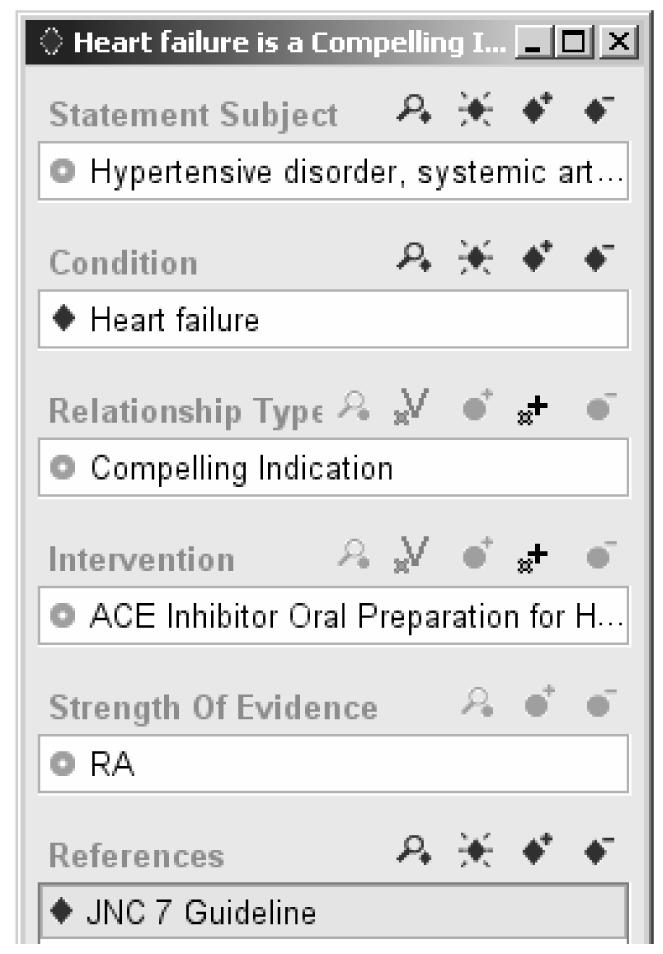
A simplified instance of a SAGE Evidence Statement, encoding the statement "In the context of treatment of hypertension, presence of heart failure is a compelling reason for the use of ACE inhibitor, with strength of evidence RA from JNC 7 guideline," where RA is the code used in JNC 7 for statements based on randomized controlled trials.
These Evidence Statements are declarative in the sense that there are no implied actions or prescribed behavior. Like records in a database, they can be queried using a structured query language. Unlike a database record, however, using these Evidence Statements to provide guideline-directed care requires that conditions (such as presence of heart failure) be evaluated in terms of available patient data. To solve this problem, the SAGE project defines a query template that allows the specification of queries that return instances of Evidence Statements for which the patient condition evaluates to either true or false. The template follows the structure of the Evidence Statement very closely. Figure 4 shows a query to find all instances of Evidence Statement for which the following are true:
Figure 4.
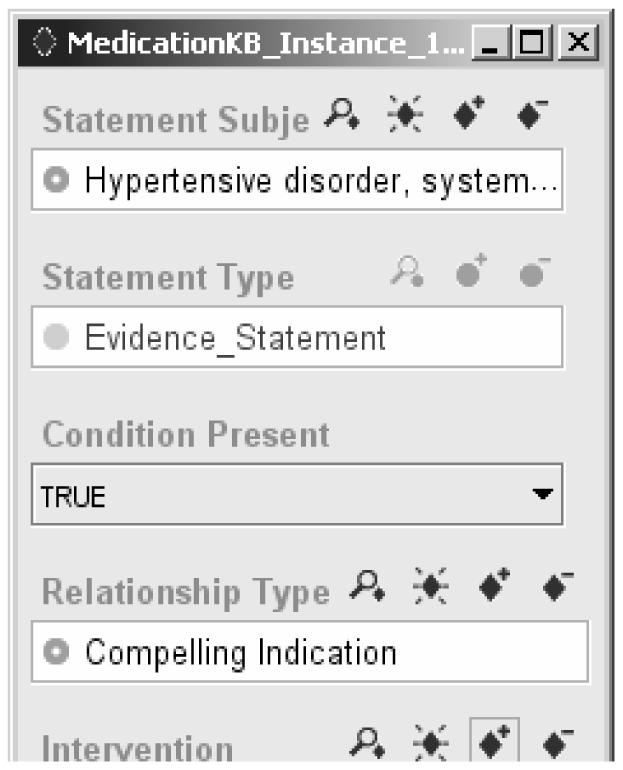
A SAGE query for instances of Evidence Statement for which, in the context of managing hypertension, the patient condition that is a compelling indication of ACE inhibitor evaluates to true.
The statement applies to the context of managing Hypertension (as seen in the statement subject).
The condition (such as the Heart Failure shown in Figure 3) is a condition the patient actually has (Condition Present = TRUE).
The relationship type to the intervention is Compelling Indication or any relationship subsumed by Compelling Indication.
The directed intervention is ACE inhibitor Oral Preparation or any drug preparation subsumed by it.
Thus, instead of using a complex, albeit expressive, logical language as EON does, the SAGE project uses a simple query template that significantly eases the encoding burden. These queries are incorporated into decision criteria for evaluating appropriate guideline-directed decisions and actions.
RESULTS
The impact of introducing declarative statements in the EON/ATHENA and SAGE decision-support systems can be seen in three ways: reduction in the effort required to develop and maintain the guideline knowledge base, the use of the declarative statements for multiple purposes, and the ability to provide nuanced decision support to clinicians.
First, instead of modeling guideline recommendations for the use of each anti-hypertensive agent as individual rules or algorithms decision criteria can be formulated as more abstract strategies, resulting in a more compact knowledge base that is easier to maintain. For example, one of the ATHENA criteria for proposing drug substitution is: if blood pressure is within targets, and there exists a prescribed antihypertensive agent that has no specific indication, and there exists a drug class that is compellingly indicated and not already prescribed or contraindicated, then consider substitution. The ATHENA knowledge base models thirteen classes of anti-hypertensive drugs for which there are 163 declarative statements linking the drug classes to 82 distinct patient situations. Modeling the usage of these drug classes individually according to their specific properties would result in vast explosion of the number of special cases.†
The separation of declarative statements from the algorithmic component of the ATHENA knowledge base that generates specific recommendations makes the knowledge base easier to maintain. Because the decision criteria of the hypertension guideline are expressed using more abstract language, when clinicians updated the knowledge base to conform to the recommendations of the latest VA guideline and JNC 7, they made only minimal modifications to the clinical algorithm (the addition of one scenario with a single decision and action). In contrast, 34 changes in the declarative statements about relationships between drugs and patient conditions (21 additions, 10 deletions, and 3 modifications) were made.
The SAGE project reported similar ease in creating and maintaining the declarative Evidence Statements. Clinicians on the project decided to select and consolidate, from multiple sources (such as JNC 7, Epocrates, and MICROMEDEX®), the drug information clinically relevant to the management of patients who have community-acquired pneumonia or both diabetes and hypertension. After they researched and reviewed the information to be encoded, one clinician informatician (KMH) was able to encode 395 instances of Evidence Statements in approximately one day. A second clinician informatician (JG) reviewed them without finding any encoding errors.
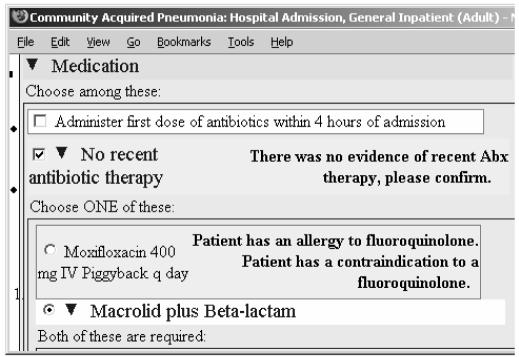
A second consequence of introducing the declarative statements is that they are available for multiple uses. As the drug substitution decision criterion described earlier illustrates, a DSS can apply these declarative statements to generate therapy recommendations. In addition, the result of querying for and evaluating these statements can be displayed wherever such information is useful. The SAGE DSS. for example, embeds queries, such as the one shown in Figure 4, in the alert and explanation texts of order sets. These queries are evaluated to generate patient-specific annotations for order sets used to deliver guideline-based recommendations to clinicians (Figure 5).
Figure 5.
Part of an order set for community acquired pneumonia where the medication order Moxifloxacin 400 mg IV Piggyback q day has allergy and relative contraindication annotations that the SAGE DSS generates by querying for and applying declarative statements in the knowledge base to the patient case.
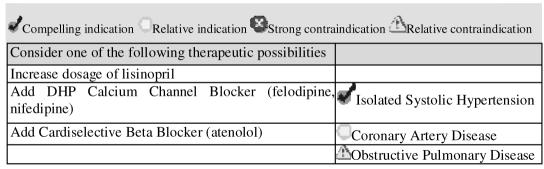
Finally, the declarative statements, when evaluated for a specific patient by the decision-support system, can present pros and cons of alternative recommended therapy choices. ATHENA DSS, for example, instead of trying to determine a best drug choice, displays patient-specific indications and contraindications as part of the explanation for choosing among multiple drugs to add or substitute (Figure 6).8 Such explanation helps clinicians to make informed choice among the alternatives suggested by the system.
Figure 6.
Part of ATHENA's drug recommendation showing patient-specific declarative statements as the rationale for the recommended interventions.
DISCUSSION
The experience of using declarative statements in the EON/ATHENA and SAGE projects reported in this paper should be seen as work that points to new directions for the standardization of guideline representation and for research on guideline modeling.
A number of researchers have proposed the standardization of the representation format of computer-interpretable guidelines (CIGs) as a strategy to promote their adoption.9 Standardization allows sharing of CIGs across multiple institutions and may attract commercial vendors to implement such CIGs. The Clinical Guideline Special Interest Group (CG SIG) was formed at Health Level Seven (HL7) with the explicit goal of establishing such a standard. Members of the CG SIG had examined a number of implemented guideline representation formalisms but have yet to propose a standard format. The syntax and semantics of the control structures and process specification languages used in different CIG formalisms are quite variable and may not be sufficiently well-defined for standardization. Furthermore, there is not yet a consensus on the appropriate structure of a CIG. For example, the InterMed project implements clinical algorithms in a top-down manner, starting with conceptual algorithms as the top layer and refining them into computable and then deployed levels.10 The SAGE project, on the other hand, specifies, at top level, actions that a decision-support system performs in reaction to opportunities to provide assistance in the clinical workflow, and encodes the medical logic of guidelines as re-usable subguidelines.11
Declarative statements such as those described in this paper, in contrast to current guideline models, express relatively simple relationships. It should be easier to reach consensus on the appropriate format and on the possible types of relationships. Such consensus would allow these declarative statements, like patient data, to be shared and used by different guideline modeling and execution systems. Furthermore, these declarative relationships are consistent with the Act Relationships in HL7’s Reference Information Model. GELLO, HL7’s object-oriented expression language for specifying decision criteria, formulae, and constraints,12 is already capable of formulating the necessary definitions of patient conditions, although it lacks an evaluate operator that, as we saw in SAGE’s query template, allows the evaluation of Boolean criteria with available patient information as part of a query on a knowledge base.
Creation of evidence statements employing national standard vocabularies such as SNOMED-CT also furthers the goal of the National Health Information Infrastructure to achieve interoperable decision support. Published as simple n-ary relationships with standard concept references, these knowledge constructs provide sharable extensions to vocabulary standards which may be employed on multiple decision support platforms. They may, in fact, be authored and maintained separately from CPGs themselves as part of an organization’s knowledge management strategy.
Our projects have demonstrated the utility of modeling, as declarative statements, significant portions of a few guidelines for chronic disease management, for community-acquired pneumonia, and for immunization. It remains to be seen to what extent this approach can be generalized to other types of guidelines. Our experiences also suggest that these small-grained declarative statements are a good way to combine knowledge that is explicitly spelled out in the guideline and knowledge that is not. In both projects, many of the relationships added to the guideline knowledge bases were not directly derived from CPGs but were gleaned from literature. The same approach also holds promise for combining declarative statements from multiple guidelines when managing patients who have several co-morbidities. While we expect guideline authors to formulate consensus recommendations that are internally consistent, this representation of evidence statements can potentially be used to encode and present conflicting recommendations from different studies with appropriate attribution of sources.
Acknowledgement
The SAGE project was supported in part by grant 70NANB1H3049 of the NIST Advanced Technology Program. The ATHENA project was supported in part by VA HSR&D CPG-97-006, CPI-275, and RCD-96301. The EON project was supported by NIH LM05708. Views expressed are those of the authors and not those of the Dept of VA and other funding agencies.
Footnotes
The eight relationships are Compelling indication, absolute contraindication, relative indication, relative contraindication, drug partner, drug partner to avoid, side effect, and complicating factor (whose presence signals that the usage of the drug class is beyond the scope of the guideline knowledge base).
For example, seven of the thirteen ATHENA drug classes have compelling indications. Thus, if we were to model the substitution criterion stated in the text as individual rules comparing current medications and their possible substitutes, we would need 91 (13 x 7) rules, with each of them having a complex if part involving conditions that represent indications and contraindications of the drugs involved.
REFERENCES
- 1.de Clercq PA, Blom J, Korsten HH, A H. Approaches for creating computer-interpretable guidelines that facilitate decision-support: A review. Artif Intell Med. 2004;31(1):1–27. doi: 10.1016/j.artmed.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Gennari JH, Musen MA, Fergerson RW, et al. The evolution of Protégé: An environment for knowledge-based systems development. Int J Hum Comput Stud. 2003;58(1):89–123. [Google Scholar]
- 3.Quaglini S, Dazzi L, Gatti L, Stefanelli M, Fassino C, Tondini C. Supporting tools for guideline development and dissemination. Artif Intell Med. 1998;14(1–2):119–137. doi: 10.1016/s0933-3657(98)00019-0. [DOI] [PubMed] [Google Scholar]
- 4.Shiffman RN, Michel G, Essaihi A, Thornquist E. Bridging the guideline implementation gap: A systematic, document-centered approach to guideline implementation. J Am Med Inform Assoc. 2004;11:418–426. doi: 10.1197/jamia.M1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein MK, Hoffman BB, Coleman RW, et al. Implementing clinical practice guidelines while taking account of changing evidence: ATHENA, an easily modifiable decision-support system for management of hypertension in primary care. Proc AMIA Symp. 2000:280–284. [PMC free article] [PubMed] [Google Scholar]
- 6.Tu SW, Musen MA. A flexible approach to guideline modeling. Proc AMIA Symp. 1999:420–424. [PMC free article] [PubMed] [Google Scholar]
- 7.Tu SW, Musen MA. Modeling data and knowledge in the EON guideline architecture. MedInfo. 2001:280–284. [PubMed] [Google Scholar]
- 8.Shankar RD, Martins SB, Tu SW, Goldstein MK, Musen MA. Building an explanation function for a hypertension decision-support system. Proc MedInfo. 2001:538–542. [PubMed] [Google Scholar]
- 9.Boxwala AA, Peleg M, Tu SW, et al. GLIF3: A representation format for sharable computer-interpretable clinical practice guidelines. J Biomed Inform. 2004;37(3):147–161. doi: 10.1016/j.jbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Peleg M, Boxwala AA, Tu SW, et al. The Intermed approach to sharable computer-interpretable guidelines: A review. J Am Med Inform Assoc. 2004;11(1):1–10. doi: 10.1197/jamia.M1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu SW, Musen MA, Shankar R, et al. Modeling guidelines for integration into clinical workflow. Proc Medinfo. 2004:174–178. [PubMed] [Google Scholar]
- 12.Sordo M, Boxwala A, Ogunyemi O, Greenes R. Description and status update on GELLO: A proposed standardized object-oriented expression language for clinical decision support. Proc Medinfo. 2004:164–168. [PubMed] [Google Scholar]