Abstract
The encoding of clinical practice guidelines into machine operable representations poses numerous challenges and will require considerable human intervention for the foreseeable future. To assist and potentially speed up this process, we have developed an incremental approach to guideline encoding which begins with the annotation of the original guideline text using markup techniques. A modular and flexible sequence of subtasks results in increasingly inter-operable representations while maintaining the connections to all prior source representations and supporting knowledge. To reduce the encoding bottleneck we also employ a number of machine-assisted learning and prediction techniques within a knowledge-based software environment. Promising results with a straightforward incremental learning algorithm illustrate the feasibility of such an approach.
Introduction
Numerous efforts are under way to develop standards for sharable guideline representations. Proposed approaches form a spectrum ranging from document-centric, text-based representations to more machine-oriented ones. These representations share many underlying features and challenges. The encoding of guidelines can be an arduous task requiring skilled human expertise and thus presents a bottleneck.
Interoperability with clinical systems requires the identification of specific recommendations along with their conditional criteria. These elements need to be encoded using a unified terminology, such as the Unified Medical Language System (UMLS) [1], to allow the delivery of relevant recommendations within clinical systems such as Electronic Medical Records (EMRs). Since all of these are difficult tasks, they are prime candidates for the employment of intelligent, knowledge-based tools.
Our approach involves the progressive enhancement of text-based guidelines by annotating them with the information required for interoperability. We have concentrated on the identification and encoding of conditional information associated with recommendations. Assuming a human expert must play the central role in the encoding process, we have explored several techniques for providing machine assistance during this process. These include offering encoding suggestions and the means for improving them over time.
Encoding Methodologies
Since the traditional and most common representation for clinical guidelines is natural language text, now often available in the HyperText Markup Language (HTML), this format is a natural starting point for the encoding process. But while great strides have been made in the fields of natural language processing and knowledge representation, the subtle nature of medical information is likely to require human interpretation. The contents of clinical practice guidelines, the result of a carefully reviewed consensus process, can be particularly difficult to interpret outside of the original natural language context. Thus, it can be advantageous for guideline representations to maintain links to these original documents.
We have proposed a markup approach [2] to produce a representation compliant with Extensible HyperText Markup Language (XHTML) and Extensible Markup Language (XML) standards of the World Wide Web Consortium (w3c.org). This approach retains the original guideline text while providing sufficient information for simple decision support systems and can also be used as an intermediate step toward increasingly sophisticated, machine-oriented, interoperable representations. We have defined a core set of markup tags constituting a Hypertext Guideline Markup Language (HGML), which are likely to be important elements of any interoperable guideline standard. Intended to supplement the tags making up XHTML, the HGML tags can be seen as a small but integral subset of the XML-based GEM [3] representation, and can be transformed to analogs in numerous other systems that allow for situation-action rules, such as the Arden Syntax [4].
Several other researchers have advocated a multistage encoding process to successively transform guideline documents from text to more computer-oriented representations. For example, Svatek [5] has proposed a five step process that adds increasingly rich semantic tags and results in a completely computational representation that combines situation-action rules, with more complex knowledge structures that allow for branching logic, state transitions and plans. Shahar and Shalom [6] envisioning a multi-stage process, have developed tools to convert text to an intermediate semi-structured text format before conversion into the Asbru or other representations [7]. Even though most approaches that begin with text use editing tools to construct relatively abstract XML representations, such as GEM [3], Asbru [7] and NewGuide [8], document-centric approaches are also being developed around original text in PDF format [9].
Automated Assistance
While the standards for integration with clinical systems are still under development, the UMLS is central to many of them. Friedman [10] has demonstrated the ability to automatically map clinical terms from reports to UMLS codes with performance comparable to human experts. The National Library of Medicine (NLM) MetaMap utility [11] has also been shown effective, primarily for patient records [12].
Attempts to automate guideline encoding include the work of Shahar [13] and Georg[14]. Shahar’s system aids in the entry of clinical temporal-abstraction knowledge as part of an encoding tool, and has demonstrated improved efficiency over manual entry. Georg describes a system in a setting similar to ours that identifies conditions and recommendations in sentences that have deontic verbs, such as “should”.
Methods
We have previously developed a markup language, HGML [2], for labeling the individual recommendations and the associated conditional logic within natural language text, to allow for interoperability while preserving the original content. Our present system attempts to assist with this process by automatically annotating these documents with encoding suggestions.
As a test case, we performed a manual HGML encoding of the JNC7 guideline for hypertension [15] identifying 70 top-level conditional expressions containing 128 (105 unique) patient characteristics and 89 associated top-level recommendations containing 74 (57 unique) actions. For this study, our concern was to predict the labels of the innermost terms, the patient characteristics and actions, rather than the top-level expressions that include complete sentences or logical connectives. Furthermore, we did not consider didactic and background content preceding the encoded sections of the guideline (before the “Ambulatory BP Monitoring” section).
Our system first applies MetaMap to the guideline text to identify possible mappings of terms to UMLS terminology and concepts. For example, consider the statement “In ((asymptomatic individuals) with (demonstrable ventricular dysfunction)), ACE inhibitors and [beta]-blockers are recommended.” Here, parentheses indicate the nested conditional expressions and underlined phrases indicate recommendations identified by a human. MetaMap identifies 5 phrases with 7 concepts [and their corresponding semantic types, shown in square brackets]:
Asymptomatic [Finding], Individual [Human],
Demonstrable [Functional Concept],
Ventricular Dysfunction [Pathologic Function],
ACE Inhibitors [Pharmacologic Substance],
Beta Blockers [Pharmacologic Substance],
Recommending [Therapeutic|Preventive Procedure].
Initial Heuristics
The system can then use the semantic types that correspond to each concept to predict the labels that appear in the human encoding. For example, Findings, and Pathologic Functions often appear as conditional criteria, while Pharmacologic Substances often appear in recommendations.
To develop an initial set of heuristics, we first divided all of the semantic types of the UMLS into mutually exclusive groups that might predict each of the HGML condition and recommendation elements.
Condition element ←
Clinical Attribute, Disease or Syndrome, Event, Finding, Sign or Symptom, etc.
Recommendation element ←
Educational Activity, Pharmacologic Substance, Therapeutic or Preventive Procedure, etc.
However, we know from experience that a Pharmacologic Substance may also appear in conditional criteria, following phrases such as “patients taking”. Thus, in general, the role cannot be determined by semantic type alone, but requires the consideration of more specific context as well.
Strategies for Learning from Experience
From a rule-based perspective, machine learning of a classification problem involves the use of examples to refine a set of rules to improve predictive accuracy. Numerous methods exist, each having trade-offs in precision and recall. We performed several experiments to assess the ability to differentiate between condition and recommendation terms using MetaMap results as features.
Our first approach was to refine the initial rule set by remembering the specific terms that are misclassified. This method, being highly specific, is not likely to generalize well to unseen cases, but is expected to demonstrate maximum improvement in accuracy. To simulate incremental feedback during the encoding process, the algorithm sequentially considers each term labeled as a condition or recommendation in the human encoding. When the predicted and observed labels differ, system predicts the first observed label for all future instances of the term. False positive predictions result in suppression of future predictions for the term until positive labels are observed. This helps to reduce false positives before encodings of a term are observed while favoring recall over reducing false positives afterward.
More generalizable results can be achieved by refining the rule-set at the level of semantic types. To explore the power of semantic types as contextual features for classification, we focused on the specific problem of differentiating 132 condition terms from the other terms within the scope of 61 recommendation segments in the human encoding of the guidelines. We proceeded using a batch process without cross-validation to determine whether the given features are sufficient to solve this problem, and to assess the trade-offs of different types of rules in learning performance. We began with an initial set of (17) semantic types sufficient to achieve maximum recall of terms labeled as conditions in the human encoding. We then considered the difference in accuracy achieved by three alternate refinements of this rule-set. We defined rules by observing the semantic types of the terms adjacent to those labeled as conditions so that predicting a condition requires that the term's semantic type additionally has
either neighbor : “OR Neighbors”
both neighbors : “AND Neighbors”
both of the two adjacent neighbors: “AND 2 Neighbors”
All three of these refinement strategies make the existing rules more specific, thus preserving recall of positive examples while potentially reducing negative examples.
Results
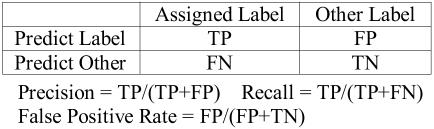
In our first step, we applied MetaMap (Rel 2.4.A, 2004_relaxed dataset) to the relevant body of the Hypertension guideline which identified 1861 terms (703 unique). We then applied our predictive algorithm, measuring accuracy before, during, and after learning. These measures are defined, conventionally, in the contingency table in Figure 1.
Figure 1.
Statistics Reported for Prediction of Human Labeled Terms
In this problem, there are several ways of measuring precision, defined as the proportion of terms whose predicted label agrees with the human label. These measures differ by the scope, and thus total number, of terms being classified. The broadest scope corresponds to the classification of labeled terms among those of the entire document. This results in the lowest measure of precision and the highest rate of false positives. We include this measure of false positive rate in our results to illustrate the difficulty of the generalized task. The narrowest scope corresponds to the classification of terms among human labeled terms only. This allows us to assess the ability to differentiate between conditions and recommendations, and is the measure of precision shown in Figure 2.
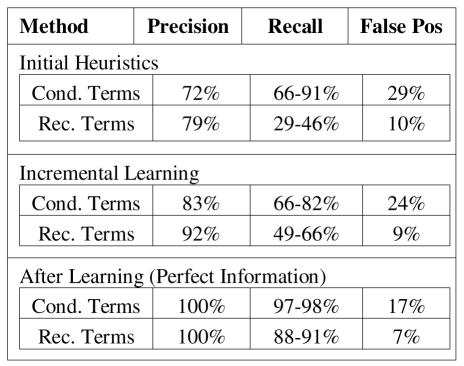
Figure 2.
Accuracy of predicted labels
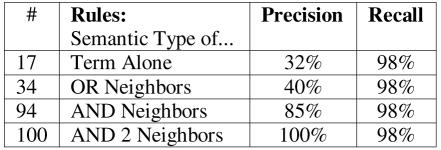
The HGML markup annotation approach also allows for nested labeling of the document, where recommendation segments may contain conditional sub-expressions or specific recommended actions. Since the identification of such recommendation segments is likely to be highly subjective, we suspect that the most useful scenario for semi-automated assistance is the identification of conditional expressions within these segments. The precision of predicting the 132 condition terms within 61 recommendation segments of the human encoded Hypertension guideline appears in Figure 3.
Figure 3.
Predicting 132 conditions within 61 recommendation segments
The measure of recall, the proportion of human labeled terms properly classified, will be the same for all of the above scopes. However, this measure is complicated by the variable nature of the markup task. Since the manually marked-up regions that identify conditions and recommendations can be of arbitrary length or overlap one another, they will not always be in one-to-one correspondence to the MetaMap terms. For this reason, we report two measures of recall, the first at the level of MetaMap terms and the second at that of the human markup. When the human markup contains multiple MetaMap terms or overlaps them, we deem a single match sufficient for this second, more lenient measure. This allows for the imperfect match and the likelihood that human concepts contain multiple components.
We observed a wider variety of semantic types in recommendations, which helps to account for the low initial recall of recommendations. Additional preliminary experiments suggest that it is possible to increase the detection of recommendation terms through the addition of a list of drugs and drug classes into our vocabulary.
When the system learns by incrementally adapting to errors, we find expected improvements in accuracy, with the notable exception of a decrease in the recall of condition terms. Upon closer inspection, we found a number of terms that were predicted as false positive conditions, when first encountered, but subsequently appeared as true positive conditions. In these cases, the initial negative feedback would result in a forgotten association that had to be re-learned later. This illustrates the need for more sophisticated uses of feedback and context.
Following the incremental learning procedure, an additional pass through the document provides insight into the ability to predict roles in this particular guideline using perfect information about the mapping of terms. We see that it is possible to predict roles with high accuracy, however mapping errors in 3 out of 132 terms limited the maximum recall to 98%. Our experiments predicting conditions within recommendation segments were based on the 17 semantic types of the learned rule-set to achieve maximum recall. We then considered three increasingly more specific rule-sets that cover the same set of positive examples. In Figure 3, we see improvements in precision as the number of clauses (#), and thus the specificity, of these rules increases.
Discussion
Our present results suggest that it is possible to provide assistance in identifying concepts and role of terms necessary to encode guidelines. In particular, the semantic types of concepts show promise in differentiating conditions from recommendations.
While our initial algorithm explores the value of semantic type as a feature alone, a comprehensive approach should also take linguistic features into account. A more syntactic approach for identifying semantic roles is taken by Georg [14], whose system parses sentences that have deontic verbs. Future approaches will most likely need to combine both syntactic and semantic features for optimal performance.
Our study considered a single representative guideline, familiar to many researchers. Transfer of learned knowledge and performance with other guidelines will depend on the similarity of the concepts and semantic types they use, and to some extent the writing style. Our methods of rule-set refinement were specifically chosen to favor improvements in accuracy over transferability to other guidelines. Ideally, for better generalization, a more sophisticated learning algorithm should strive for a minimal number of rules while maximizing accuracy. The generation and assessment of more optimal rule-sets will benefit from cross-validation with bigger training sets.
Although we cannot rely on our limited results to predict performance on other guidelines, we note the high accuracy in both precision and recall of our results. This suggests the capability to perform well for revisions of this specific guideline and potentially other similar guidelines. However, the ability to direct the encoding process by identifying likely conditions and recommendations is limited by the large number of false positive elements. This is expected due to the subjective nature of the statements selected by experts as recommendations. Thus, we feel the most useful application will be for predicting conditional expressions within manually identified recommendations segments.
Further experience encoding more guidelines is required to determine whether assistance helps or hinders the encoding process. It is presently unclear whether the resulting suggestions are likely to be reasonable and relevant, or unreasonable and overwhelming. However, the promise of an intelligent, adaptive tool is to make the encoding process easier as more experience is accrued.
Acknowledgments
We thank several reviewers for their suggestions.
References
- 1.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993:281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagerty CG, Pickens D, Kulikowski C, Sonnenberg F. HGML: A Hypertext Guideline Markup Language. In: Overhage JM, editor. Proceedings of the AMIA Symposium. 2000. pp. 325–9. [PMC free article] [PubMed] [Google Scholar]
- 3.Shiffman RN, Karras BT, Agrawal A, Chen R, Marenco L, Nath S. GEM: a proposal for a more comprehensive guideline document model using XML. J Am Med Inform Assoc. 2000:488–98. doi: 10.1136/jamia.2000.0070488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton PD, Pryor TA, Wigertz OB, Hripcsak G. Issues and structures for sharing knowledge among decision-making systems: The 1989 Arden Homestead Retreat. In: Kingsland LC, editor. Proceedings of the Thirteenth Annual Symposium on Computer Applications in Medical Care. New York: IEEE Computer Society Press; 1989. pp. 116–21. [Google Scholar]
- 5.Svatek V, Kroupa T, Ruzicka M. First European Workshop on Computer-based Support for Clinical Guidelines and Protocols. Leipzig 2000: IOS Press; 2001. Guide-X - a Step-by-step, Markup-Based Approach to Guideline Formalisation; pp. 97–114. [Google Scholar]
- 6.Shalom E, Shahar Y. A Graphical Framework for Specification of Clinical Guidelines at Multiple Representation Levels. Proceedings AMIA Annual Symposium. 2005:679–683. [PMC free article] [PubMed] [Google Scholar]
- 7.Kosara R, Miksch S, Seyfang A, Votruba P. Integrated Design and Process Technology, IDPT-2002. Jun, 2002. Tools for Acquiring Clinical Guidelines in Asbru. [Google Scholar]
- 8.Ciccarese P, Kumar A, Quaglini S. New-Guide: a new approach to representing clinical practice guidelines. Advances in Clinical Knowledge Management-5. 2002:15–18. [Google Scholar]
- 9.Eriksson H, Tu S, Musen M. Semantic Clinical Guideline Documents. Proceedings of the AMIA Annual Symposium. 2005:236–240. [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated Encoding of Clinical Documents Based on Natural Language Processing. J Am Med Inform Assoc. 2004;11:392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronson A. MetaMap: Mapping Text to the UMLS Metathesaurus. [monograph on the Internet] National Library of Medicine. 1996. [cited 2005 March 3]. Available from: http://ii.nlm.nih.gov/resources/metamap.pdf.
- 12.Evans DA, Brownlow ND, Hersh WR, Campbell EM. Automating concept identification in the electronic medical record: an experiment in extracting dosage information. Proceedings of the American Medical Informatics Association Annual Symposium. 1996:388–92. [PMC free article] [PubMed] [Google Scholar]
- 13.Shahar Y, Chen H, Stites DP, Basso L, Kaizer H, Wilson DM, Musen MA. Semiautomated Entry of Clinical Temporal-Abstraction Knowledge. J Am Med Inform Assoc. 1999;6:494–511. doi: 10.1136/jamia.1999.0060494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georg G, Jaulent M-C. An Environment for Document Engineering of Clinical Guidelines. Proceedings of the AMIA Annual Conference. 2005:276–280. [PMC free article] [PubMed] [Google Scholar]
- 15.Chobanian AV, et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure, The JNC 7 Report. J Am Med Assoc. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]