Abstract
Hostile physiological environments such as hypoxia and acidic extracellular pH, which exist in solid tumors, may promote invasion and metastasis through inflammatory responses and formation of eicosanoids. Here, we have investigated the effects of the anti-inflammatory agent indomethacin on the invasion and metabolism of the human breast cancer cell line MDA-MB-435 in Dulbecco's Modified Eagles (DME)-based or Roswell Park Memorial Institute (RPMI)-based cell medium, using a magnetic resonance-compatible invasion assay. Indomethacin treatment significantly reduced the invasion of MDA-MB-435 cells independent of the culture and perfusion conditions examined. Significant changes were detected in levels of intracellular choline phospholipid metabolites and in triglyceride (TG) concentrations of these cells, depending on indomethacin treatment and basal cell medium used. Additionally, genetic profiling of breast cancer cells, grown and treated with low-dose indomethacin in cell culture using an RPMI-based medium, revealed the upregulation of several genes implicating cyclooxygenase-independent targets of indomethacin. These data confirm the ability of an anti-inflammatory agent to reduce breast cancer invasion and demonstrate, depending on cell culture and perfusion conditions, that the indomethacin-induced decrease in invasion is associated with changes in choline phospholipid metabolism, TG metabolism, and gene expression.
Keywords: Breast cancer, indomethacin, invasion, phospholipid metabolism, magnetic resonance (MR)
Introduction
Solid tumors, including breast cancers, are often characterized by hypoxia, extracellular acidosis, and nutrient deprivation [1]. Similar to wounds, these characteristics of the tumor microenvironment can evoke an inflammatory response [2–4]. Not surprisingly, solid tumors frequently exhibit increased levels of pro-inflammatory molecules, such as prostaglandins, that are secreted by tumor cells, tumor-associated stromal cells, and inflammatory cells [5,6]. These prostaglandins can impact cancer cell motility, invasion, vascularization, and metastatic dissemination [6–8]. Prostaglandins are synthesized from arachidonic acid by the action of cyclooxygenases (COXs) [5,6], and increased expression of COX-2 is also frequently observed in tumors [7]. Nonsteroidal anti-inflammatory drugs (NSAIDs) such as indomethacin reduce inflammation by inhibiting COX activity [9], and clinical findings of more than a decade have established that NSAIDs can prevent an array of cancers, including estrogen receptor-positive breast cancer [10]. Free arachidonic acid is formed from the breakdown of membrane phospholipids by phospholipases, such as phospholipases A2, C, and D, in response to mechanical, chemical, and physical stimuli [11,12]. Therefore, prostaglandin and eicosanoid biosynthesis and membrane choline phospholipid metabolism are closely coupled [11,12]. We have previously observed that increased intracellular phosphocholine (PC) and total choline [tCho; free choline + PC + glycerophosphocholine (GPC)] were associated with increased malignancy [13,14]. We also observed that treatment of breast cancer cells with indomethacin significantly reduced intracellular PC levels and increased intracellular GPC levels toward levels typical of less malignant human mammary epithelial cells (HMECs) [15–17]. Recently, we also investigated the contribution of the anabolic and catabolic pathways to the altered metabolic phospholipid profile following treatment of MDA-MB-231 breast cancer cells and nonmalignant immortalized MCF-12A cells with 300 µM indomethacin for 3 hours [18]. These studies revealed that short-term, high-dose indomethacin treatment significantly altered the genetic profile of treated cells [18].
To further understand the role of COX and choline phospholipid metabolism in breast cancer invasion, here, we investigated the effect of long-term low-dose indomethacin on breast cancer cell invasion using a magnetic resonance (MR)-compatible cell perfusion assay, the Metabolic Boyden Chamber (MBC), to dynamically track invasion and metabolism. The ability of the MBC assay to measure invasion and cellular metabolism under controlled environmental conditions in long-term, MR experiments has been previously validated using three prostate and three breast cancer cell lines preselected for differences in invasive behavior [19,20]. This assay employs extracellular matrix (ECM) gel, which is derived from Engelbreth-Holm-Swarm murine sarcoma and is composed primarily of laminin, collagen type IV, heparan sulfate proteoglycan, and entactin as a barrier for cellular invasion (i.e., cellular migration through and proteolytic degradation of ECM).
The incorporation of slow-release indomethacin pellets into the assay resulted in a significant reduction in ECM gel degradation by cancer cells compared to controls. Interestingly, we observed that perfusing cells with Roswell Park Memorial Institute (RPMI) or DME as basal cell culture medium also altered the ability of these cells to degrade ECM. Invasion and ECM degradation were enhanced for cancer cells maintained in an RPMI-based culture medium compared to cells maintained in a DME-based culture medium. Consistent with our previous observations, indomethacin treatment induced changes in choline phospholipid metabolites to resemble levels more typical of nonmalignant cells. Additionally, intracellular mobile lipid levels increased as a result of indomethacin treatment.
In addition to nonspecific COX inhibition, indomethacin has several other effects. It can bind and activate peroxisome proliferator-activated receptor gamma (PPARγ), stimulate NAG-1 expression and proapoptotic activity, and cause apoptosis by releasing cytochrome c and apoptosis-inducing factors and by caspase activation [5,7]. In addition, it has been shown to activate carbonic anhydrase I and carbonic anhydrase II isozymes in a dose-dependent manner, to downregulate β-catenin/TCF signaling in colorectal cancer cells, and to act as a mitochondrial uncoupler [5,7].
To further understand the mechanisms underlying the functional changes in invasion and metabolism detected in our studies, we used oligonucleotide microarray technology to determine changes in gene expression levels induced by indomethacin treatment in RPMI-based cell culture. Consistent with previous studies [21], we found that GDF15, a divergent member of the TGF-β superfamily, was upregulated by indomethacin treatment. The GDF15 gene encodes for a protein that is involved in differentiation, apoptosis, and other biologic functions through autocrine or paracrine signaling [21]. The expression of GDF15 was shown to be COX-independent and p53-independent in several cancer cell lines [22]. In addition to GD15, changes in several COX-independent genes were identified in microarray analysis, suggesting that, in addition to COX inhibition, other mechanisms may play a role in the reduced invasion and altered metabolism observed in this study. Our data support the use of anti-inflammatory agents in breast cancer management and prevention.
Materials and Methods
Cells and Cell Culture
Experiments were performed with the human breast cancer cell line MDA-MB-435, which was originally isolated from the pleural effusion of a 31-year-old female with metastatic ductal adenocarcinoma of the breast and established in cell culture [23]. MDA-MB-435 breast cancer cells were cultured in plastic tissue culture at 5% CO2 in air, 37°C, and 90% humidity using two different cell culture media: DME plus supplements (DMEC) and RPMI 1640 plus supplements (RPMIC). The cell culture medium DMEC was composed of DME with 25 mM glucose and 4 mM l-glutamine but without sodium pyruvate (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 9% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO), 90 U/ml penicillin (Invitrogen Corp., Grand Island, NY), and 90 µg/ml streptomycin (Invitrogen Corp.); RPMIC consisted of RPMI 1640 (Sigma-Aldrich) supplemented with 9% FBS, 90 U/ml penicillin, and 90 µg/ml streptomycin.
Three days before the MR experiments, MDA-MB-435 cells were seeded on Biosilon (Nunc, Denmark) beads at a cell density of 3 x 106 cells per 0.5 ml of microcarriers in non-cell culture Petri dishes (Labtec, Nunc, Denmark) and grown adherently to approximately 80% to 90% confluency using either DMEC or RPMIC. MDA-MB-435 cells on Biosilon beads were counted at the beginning and at the end of the MR experiments, as described previously [24]. Briefly, approximately 0.5 ml of cell-covered beads was incubated in 1 ml of 0.1 M citric acid containing 0.1% (wt/vol) crystal violet for 1 hour at 37°C, thus lysing the cells and releasing the nuclei. Subsequently, the nuclei were diluted and counted with a hemocytometer.
For GeneChip (Affymetrix, Santa Clara, CA) microarray experiments, 3 x 106 MDA-MB-435 cells were seeded per 100-mm cell culture dish using 15 ml of RPMIC. Following attachment overnight, the cell culture medium was exchanged. Cells from one treatment dish containing one 21-day-release biodegradable indomethacin pellet (0.5 mg/pellet, MW indomethacin = 357.79 g/mol; Innovative Research of America, Sarasota, FL) and from one control dish were used to isolate RNA. The dishes were placed at 5% CO2 in air, 37°C, and 90% humidity for 48 hours. A single pellet was used to achieve an indomethacin concentration comparable to that of the cell perfusion system. Two independent experiments were performed.
MBC Assay
Details of MBC assay, which was developed to noninvasively study cancer cell invasion and cancer cell metabolism, have been previously described in Pilatus et al. [25]. Modifications incorporated for the current study are described here.
MR-compatible cell perfusion system
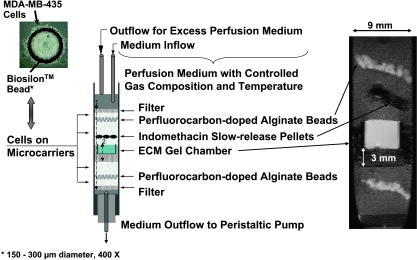
A schematic of a sample preparation is shown in Figure 1. The sample consisted of different layers containing filter material, cellcovered beads, a home-built chamber filled with ECM gel (Sigma-Aldrich), perfluorocarbon-doped alginate beads, and indomethacin pellets in treatment experiments.
Figure 1.
Schematic display of the sample structure (center), which can be identified clearly in the representative T1-weighted 1H MR image on the right. The photograph on the top left shows a representative Biosilon bead covered with MDA-MB-435 cells. Control experiments were performed without pellets or unloaded pellets. In experiments with indomethacin treatment, two or three biodegradable 21-day slow-release pellets were added to the upper cell layer approximately 4 mm above the ECM gel. The released indomethacin concentrations calculated for the total medium volume of approximately 420 ml in the perfusion system were ∼ 0.48 µmol/(l x 24 hours) for three pellets and - 0.32 µmol/(l x 24 hours) for two pellets of indomethacin. The two arrows, marked 1 and 2, reflect the region integrated to obtain Ip(t) and Ip,7 mm(t), as defined in Eq. (1). The invasion index reflects a composite of cell migration and ECM degradation.
The home-built chamber consisted of an outer chamber made from porous filter material (polyethylene; Small Parts, Inc., Miami Lakes, FL) and an inner chamber composed of a polycarbonate membrane (Millipore, Bedford, MA) fixed to a Delrin ring (McMater-Carr Supply Company, Chicago, IL). Cold liquid ECM gel (100 µl, 8.8 mg/ml protein, 4°C) was pipetted into this chamber and placed at 37°C to polymerize and form a well-defined layer (Figure 1). Cell-covered beads were layered around the ECM gel chamber in a customized 10-mm nuclear magnetic resonance (NMR) tube (Wilmad Ltd., Buena, NJ) that had a reinforced open base to accommodate the outflow line of the cell perfusion system (Figure 1). As shown in Figure 1, layers of perfluorocarbon-doped alginate beads were interspersed in the upper and lower cancer cell layers to monitor oxygen tension in the sample using 19F MR relaxometry. In some experiments, perfluorocarbon was added to the ECM gel to measure oxygen tension in the ECM layer. The preparation of perfluorocarbon-doped alginate beads and the measurement of oxygen tension using 19F MR relaxometry have been described previously in detail by Pilatus et al. [25,26]. In treatment experiments, two or three 21-day-release biodegradable indomethacin pellets (0.5 mg/pellet) were positioned in the upper cell layer approximately 4 mm above the ECM gel (Figure 1).
The sample was perfused continuously with either DMEC plus 10 mM HEPES (DMECP) or RPMIC plus 10 mM HEPES (RPMICP) (Sigma, St. Louis, MO). The flow rate of the ∼ 410-ml recirculating perfusion medium was approximately 1 ml/min. Sample temperature was maintained at 37°C, and the oxygen tension in the sample was maintained above 10%.
MR data acquisition
The MR experiments were performed on a GE Omega 400 NMR spectrometer equipped with shielded triple-axis gradients (130 G/cm maximum). MR data were acquired using a home-built radiofrequency probe consisting of a broadband coil for 31P MR spectroscopy and a second coil tunable to 1H or 19F frequency.
The following MR data sets were acquired every 12 hours, throughout the duration of an experiment that typically lasted 3 days [25]. T1-weighted 1H MR imaging was performed to evaluate the sample preparation, to visualize the geometry of the ECM gel, and to detect changes in its integrity due to invasion and degradation (Figure 1, right panel). A one-dimensional (1D) 1H profile of intracellular water was acquired along the length of the sample (z-axis), using diffusion-weighted 1D 1H MR imaging to suppress extracellular water signal [25]. This profile, acquired with a spatial resolution of 31.25 or 62.5 µm, was used to derive a quantitative index of cell invasion.
Energy metabolites, pH, and the choline phospholipid metabolites PC and GPC were obtained from global 1D 31P NMR spectra.
Intracellular levels of tCho, total creatine (tCr; creatine + phosphocreatine), and lactate + triglyceride (LacTG) were derived from global 1D 1H MR spectra acquired with diffusion weighting and CHEmical Shift Selective (CHESS) water suppression [25]. Diffusion-weighted 1D 1H MR spectra, obtained without water suppression, were used to determine cell proliferation because the increase in slow-diffusing water, which represents intracellular water, was directly proportional to the number of cells. A cell count at the beginning and at the end of each MR experiment confirmed cell growth, indicating the absence of significant cell swelling or shrinkage. To quantify the contribution of lactate (Lac) and triglycerides (TGs) to the LacTG signal at 1.3 ppm in the global 1H MR spectra, we also acquired diffusion-weighted 1D 1H MR spectra using a spin echo-based pulse sequence with an echo time of 136 milliseconds and Lac editing [27].
Localized 1D 1H chemical shift imaging (CSI) MR spectra with and without CHESS water suppression were acquired to obtain metabolic information from 310-µm-thick slices along the z-axis of the sample [25]. Data from slice-selective 1D 19F inversion recovery experiments were used to calculate oxygen tension in slices containing perfluorocarbon, as described previously [26]. Localized 1D 1H CSI and 19F MR spectra were acquired every 24 hours.
MR data processing and analyses
Proton MR images and profiles of cellular water were processed using software provided with the spectrometer and exported for further analysis. All MR spectra were processed and analyzed using XsOsNMR, a postprocessing and quantitation package programmed in Interactive Data Language (ITT, Boulder, CO). XsOsNMR was developed and kindly provided by Dr. D. C. Shungu and X. Mao (Hatch NMR Research Center, Columbia University College of Physicians and Surgeons, New York, NY).
T1-weighted 1H MR images were zero-filled once in the phase-encoding dimension followed by two-dimensional (2D) magnitude calculation. 1D profiles of intracellular water were zero-filled once (resulting in a spatial resolution of 15.6 or 31.25 µm), multiplied by a combination of exponential and Gaussian functions in the time domain, followed by a magnitude calculation of Fourier-transformed echo.
The profiles of intracellular water were used to quantify the number of cancer cells that invaded the ECM gel and to derive an index of invasion. The invasion index I(t) at time t was calculated as follows:
| 1 |
where Ip,7 mm(t) is the integral value of the signal at time t, obtained by integrating intracellular water signal over a 7-mm region starting at the base of theECMgel chamber, and Ip(t) is the integral for the entire diffusion-weighted profile at time t. The first contact of cancer cells with the ECM gel during the loading of the sample was defined as the zero time point, and t1 defines the first MR data set acquired after loading.
After applying an exponential line-broadening factor of 15 Hz to the free induction decay (FID) of 1D 31P MR spectra and 8 Hz to the FID of 1D 1H MR spectra, the resulting FIDs were Fourier-transformed, and a zero-order phase correction was applied. Spectroscopic imaging data were processed by applying a Hamming filter in the phase-encoding dimension and an exponential line broadening of 8 Hz in the time domain, followed by 2D Fourier transformation and phase correction.
Signals of interest in the 31P and 1H MR spectra were fitted in the time domain using an algorithm combining linear least squares and nonlinear least squares fitting routines. Metabolite levels in the 31P spectra [met31P(t)] at time t were calculated as follows:
| 2 |
where met31P is the normalized relative metabolite level and Im is the integral of the metabolite signal. The pH was calculated based on the equation: pH = 6.66 + log10[(ΔPi - 0.729) / (3.22 - ΔPi)], where ΔPi = chemical shift of inorganic phosphate [28]. Metabolite levels in the 1D 1H MR spectra relative to the number of cells [met1H(t)] were calculated as:
| 3 |
where met1H is the relative metabolite concentration in the proton MR spectra, Im is the integral of the metabolite signal in the 1H MR spectra, and Icell is the integral of the corresponding unsuppressed intracellular water signal at time t, which is used to account for cell growth during the time course of the experiment. Temperature was calculated from the chemical shift difference between tCho and intracellular water in the 1H MR spectra [29,30]. Both Ip and Icell are proportional to the number of cells at time t and represent cell growth during the MR experiment, as was validated from nuclei counts of the cells on Biosilon beads at the start and at the end of the MR experiments.
Fluorine MR spectra acquired during the 19F inversion recovery experiment were processed and analyzed as described in detail by Pilatus et al. [26].
Values presented as mean ± SE were averaged over three independent experiments (n = 3) for each condition, unless otherwise stated. The Mann-Whitney U test was performed to test for statistically significant differences (P < .05) between treated and untreated conditions using JMP IN 5.1 (Thompson Learning; SAS Institute, Inc., Belmont, CA).
GeneChip Microarray Experiments
Isolation of total RNA and human genome array
Total cellular RNA of MDA-MB-435 cells was isolated using the RNeasy Mini Kit (Qiagen, Inc., Valencia, CA). Briefly, MDA-MB-435 cells were washed twice in phosphate-buffered saline. The cells were scraped in 600 µl of RLT lysis buffer containing 6 µl of β-mercaptoethanol and homogenized using QIAshredder homogenizer spin columns (Qiagen, Inc.). The samples were diluted 1:2 with 70% ethanol and applied to RNeasy mini columns to capture RNA. Following DNase digestion and washing, RNA was eluted from RNeasy mini columns and quantified, and RNA purity was evaluated.
An Affymetrix GeneChip Human Genome U133 Plus 2.0 Array was used to analyze the expression levels of ∼ 47,400 transcripts, which represent 38,500 genes. RNA labeling and microarray hybridization were performed by the Johns Hopkins Medical Institutions Microarray Core Facility (Dr. F. M. Murillo; The Johns Hopkins University School of Medicine, Baltimore, MD).
Briefly, RNA underwent quality control to confirm that all samples had optimal rRNA ratios (1:2 for 18S and 28S, respectively) and clean run patterns using an Agilent Bioanalyzer (Agilent, Santa Clara, CA). Then, 5 µg of pure total RNA were converted to single-strand cDNA using oligonucleotide probes containing 24 oligodeoxythymidylic acid plus T7 promoter as primer (Proligo LLC, Boulder, CO) and the SuperScript Choice System (Invitrogen Corp.). Following the synthesis of double-stranded cDNA and product purification by phenol-chloroform extraction, biotinylated antisense cRNA was generated by in vitro transcription using the BioArray HighYield RNA Transcript Labeling System (ENZO Life Sciences, Inc., Farmindale, NY). Then, 15 µg of biotinylated cRNA were fragmented in 100 mM Tris-acetate pH 8.2, 500 mM potassium acetate, and 150 mM magnesium acetate at 94°C for 35 minutes. Ten micrograms of fragmented cRNA were hybridized to the Affymetrix U133 Plus 2.0 GeneChip array for 16 hours at 45°C under constant rotation. Using an Affymetrix Fluidics Station 450, GeneChip arrays were washed to remove nonhybridized cRNA and incubated with streptavidin-phycoerythrin conjugate to stain hybridized biotinylated cRNA. The staining was amplified using IgG (goat) as a blocking reagent and biotinylated antistreptavidin antibody (goat) followed by a second staining with streptavidin-phycoerythrin conjugate. Fluorescence intensities were measured with an Affymetrix GeneChip Scanner 3000. Basic image analysis for quality control was performed. Fluorescence intensities were measured with an Affymetrix GeneChip Scanner 3000, and a basic image analysis for quality control was performed using the Affymetrix GeneChip Operating System 1.1.1. Two independent experiments each were run for treated and untreated MDA-MB-435 cells.
Analysis of human genome array data
Additional quality control and analysis of human genome array data were performed using the R environment (http://www.bioconductor.org/, and Ref. [31]) at the Analysis Unit of the Johns Hopkins Medical Institutions Microarray Core (Dr. Chunfa C. Jie, The Johns Hopkins University School of Medicine).
A model-based quality control assessment of the Affymetrix GeneChip array was performed, followed by a principal components analysis. RNA degradation was statistically evaluated. Human genome array data exhibited low RNA degradation and excellent image quality (data not shown).
Based on the Affymetrix perfect match and mismatch probe values, gene expression intensities were calculated by Robust Multiarray Average [32]. Probe level data processing included quantile normalization to reduce the obscurity from variation between microarrays, which might be introduced during sample preparation, manufacture, fluorescence labeling, hybridization, and/or scanning [33]. An empirical Bayes method with γ-γ modeling was applied on the resulting signal intensities to estimate the posterior probabilities of the differential expression of genes between untreated and indomethacin-treated MDA-MB-435 cells [34–36]. The list of differentially expressed genes was produced by applying a posterior probability above .5 (i.e., the posterior probability is larger than chance).
Quality control and statistical analysis were performed using bioconductor packages (http://www.bioconductor.org/).
Results
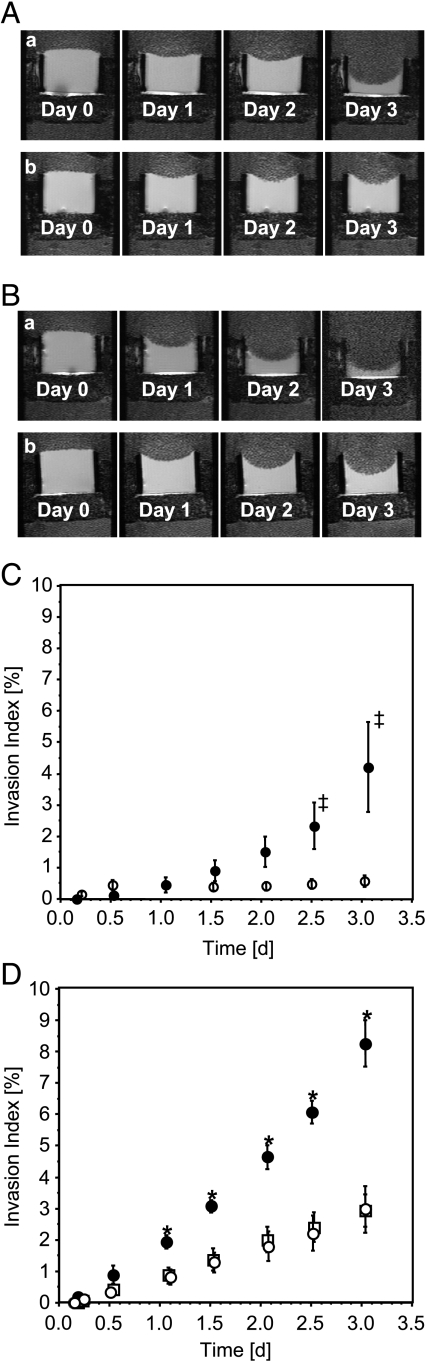
The MBC allowed us to identify the invasive and metabolic characteristics of MDA-MB-435 cells in response to low-dose indomethacin treatment dynamically, longitudinally, and noninvasively under controlled environmental conditions. Indomethacin treatment reduced the rates of ECM gel degradation, independent of the culture and perfusion media used, as evident in the representative 1H MR images shown in Figure 2, A and B. Quantitative time-dependent invasion indices I(t) demonstrated that the invasion of MDA-MB-435 breast cancer cells maintained in DMECP was almost entirely inhibited by indomethacin treatment (Figure 2C). For MDA-MB-435 cells maintained in RPMICP, indomethacin treatment significantly attenuated invasion but did not inhibit it completely; reduction of invasion was comparable for the two doses of indomethacin (Figure 2D).
Figure 2.
T1-weighted 1H MR images (A and B) and invasion indices over time (C and D) demonstrating the effect of indomethacin treatment and cell culture medium on the degradation (A and B) and invasion (A–D) of ECM gel by MDA-MB-435 breast cancer cells. Invasion includes components of cell migration and ECM degradation. (A, a) Untreated MDA-MB-435 cells cultured in DMEC and perfused in DMECP. (A, b) Indomethacin-treated (three pellets) MDA-MB-435 cells cultured in DMEC and perfused in DMECP. (B, a) Untreated MDA-MB-435 cells cultured in RPMIC and perfused in RPMICP. (B, b) Indomethacin-treated (three pellets) MDA-MB-435 cells cultured in RPMIC and perfused in RPMICP. (C) DMECP: closed circles, control, n = 4; open circles, three pellets of indomethacin, n = 3 (‡P = .16 for treated versus untreated MDA-MB-435 cells). (D) RPMICP: closed circles, control, n = 3; open circles, three pellets of indomethacin; open squares, two pellets of indomethacin (*P < .05 for treated versus untreated MDA-MB-435 cells). Values of invasion indices are presented as mean ± SE.
Untreated and indomethacin-treated MDA-MB-435 cells maintained in RPMICP exhibited a stronger invasive capability compared to MDA-MB-435 cells in DMECP under equivalent conditions.
Because the profile of intracellular water is proportional to the cell number at any given time, as was also apparent from the cell count at the beginning and at the end of the MR experiment, we calculated the fold change in cell number during the time course of the MR experiments. The cell density in the sample increased by approximately 3.6-fold for control MDA-MB-435 cells, independent of the cell culture medium used, whereas with indomethacin treatment, the cell density increased by ∼ 3-fold for cells in RPMICP and by ∼ 2.5-fold for cells in DMECP. Therefore, the difference in invasive ability under the different conditions cannot be explained by changes in cell proliferation.
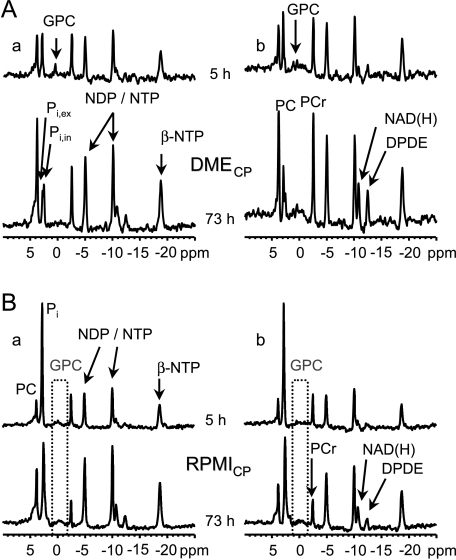
Representative 1D 31P MR spectra of indomethacin-treated and untreated MDA-MB-435 cells maintained in DMECP or RPMICP acquired after 5 and 73 hours, respectively, demonstrate the stability of pH during the time course of the experiments (Figure 3). Although the experimental setup did not permit the comparison of metabolite concentrations across separate experiments, we could track normalized relative changes (Figure 4).
Figure 3.
Representative 1D 31P MR spectra of the entire sample at the beginning and at the end of an experiment. (A, a) DMECP, control. (A, b) DMECP, globally released indomethacin concentration of 0.48 µmol/(l x 24 hours). (B, a) RPMICP, control. (B, b) RPMICP, globally released indomethacin concentration of 0.48 µmol/(l x 24 hours). Signal assignments: DPDE, diphosphodiester; NADP(H), signal consisting of NAD+, NADH, NADP+, and NADPH; PCr, phosphocreatine; NTP, nucleoside triphosphate; NDP, nucleoside diphosphate; Pi, inorganic phosphate (ex, extracellular; in, intracellular).
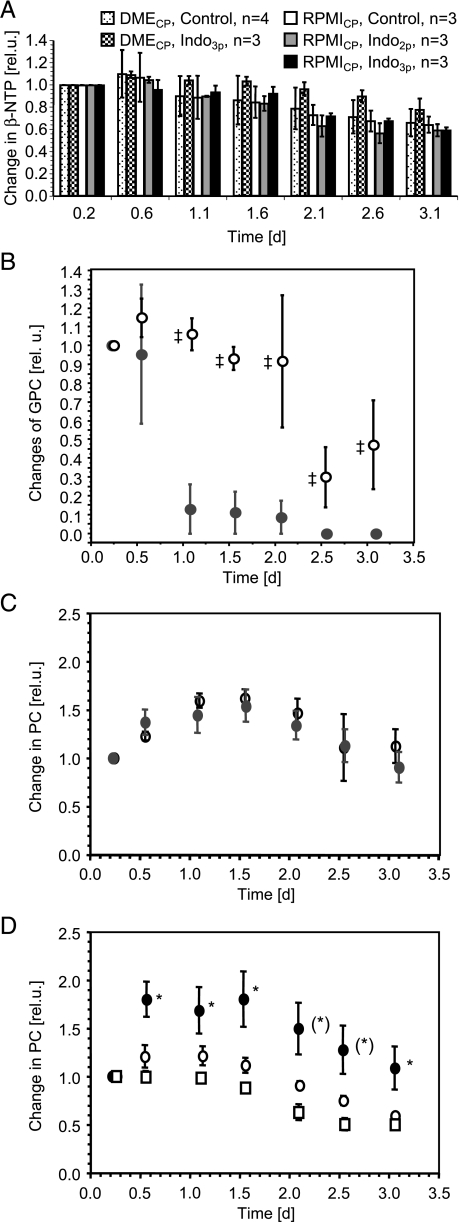
Figure 4.
Relative changes in phosphometabolites during the time course of the MR experiments. Values are presented as mean ± SE. (A) Relative changes in β-NTP for indomethacin-treated (three or two pellets of indomethacin) and untreated MDA-MB-435 cells perfused with DMECP or RPMICP. (B) Relative changes in GPC for treated (open circles, three pellets of indomethacin, n = 3) and control (closed circles, n = 4) MDA-MB-435 cells perfused with DMECP. ‡P < .05 for indomethacin-treated versus untreated MDA-MB-435 cells. (C) Relative changes in PC for control (closed circles, n = 4) and indomethacin-treated (open circles, three pellets of indomethacin, n = 3) MDA-MB-435 cells perfused with DMECP. (D) Relative changes in PC for control (closed circles, n = 3) and indomethacin-treated (open circles, three pellets of indomethacin, n = 3; open squares, two pellets of indomethacin, n = 3) MDA-MB-435 cells perfused with RPMICP. *P < .05 for indomethacin-treated versus untreated MDA-MB-435 cells. (*) Two time points where untreated MDA-MB-435 cells were significantly different from cells treated with two pellets of indomethacin, but not from cells treated with three pellets of indomethacin.
Nucleoside triphosphate levels corrected for cell number and normalized to the starting value were not significantly influenced by indomethacin treatment or change in basal medium, but tended to decrease with time to between 60% and 80% of the starting value (Figure 4A). In experiments performed with DMECP, GPC decreased within the first day to noise level in untreated MDA-MB-435 cells, whereas GPC in indomethacin-treated cells remained constant and declined to about 30% of the starting value only after 2 days (Figure 4B). In the MR experiments performed with RPMICP, the GPC signal did not rise significantly above noise in either control or indomethacin-treated breast cancer cells (Figure 3B).
Changes in intracellular PC levels over the time course of MBC experiments using DMECP were not affected by indomethacin treatment (Figure 4C). However, in MBC experiments of MDA-MB-435 cells maintained in RPMICP, indomethacin treatment significantly reduced intracellular PC levels (Figure 4D).
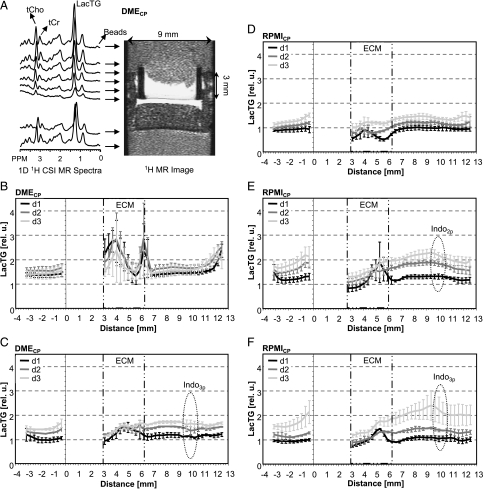
Figure 5A shows a set of 1D 1H CSI MR spectra and the corresponding region in the 1H MR image on day 2. Composite MR signals representing the intracellular concentrations of metabolites such as tCho, tCr, and LacTG in 310-µm-thick slices were identified and quantified. LacTG levels along the sample for untreated and treated MDA-MB-435 cells perfused with DMECP or RPMICP are shown in Figure 5, B–F. Figure 6A shows global cellular LacTG levels of control and treated MDA-MB-435 cells in either DMECP or RPMICP, obtained from global 1D 1H MR spectra using a STEAM-based pulse sequence.
Figure 5.
(A) Representative water-suppressed 1D 1H CSI MR spectra of 310-µm-thick slices acquired along the sample 47 hours after loading, together with the corresponding 1H MR image for MDA-MB-435 cells perfused with DMECP. (B–F) Relative levels of LacTG, along with the sample obtained for MDA-MB-435 cells. Values presented as mean ± SE are from 1D 1H CSI spectra averaged over three experiments for each condition, unless otherwise stated. (B) Untreated, DMECP, n = 4. (C) Three pellets of indomethacin, DMECP. (D) Untreated, RPMICP. (E) Two pellets of indomethacin, RPMICP. (F) Three pellets of indomethacin, RPMICP (d2; average of n = 2). The base of the ECM gel chamber was defined as 0 mm. The empty region from 0 to 3 mm represents the filter material forming the base of the ECM gel chamber. ECM = the thickness of the ECM gel at the start of the experiments; Indo = the approximate position of the indomethacin pellets in the upper cell layer at the start of the MR experiments; d1, d2, d3 = elapsed time after the loading of cells (i.e., days 1, 2, and 3, respectively).
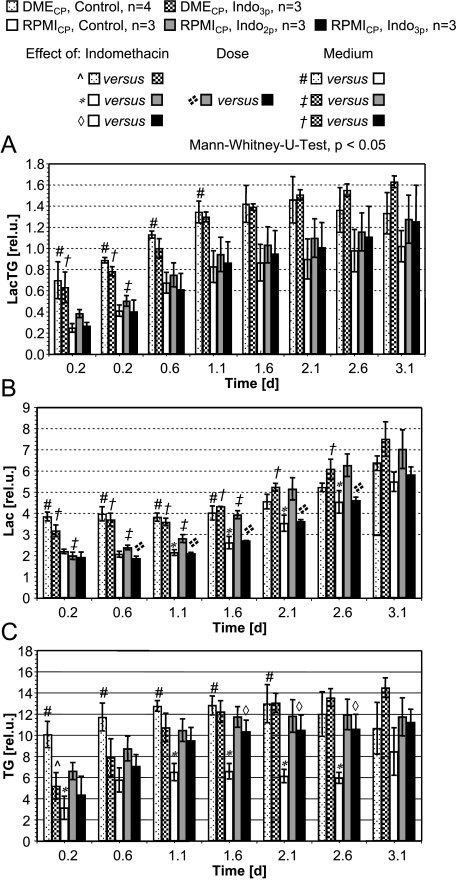
Figure 6.
(A) Relative levels of LacTG over time obtained for MDA-MB-435 cells in DMECP or RPMICP. Values presented as mean ± SE are from global 1D 1H MR spectra acquired with a STEAM-based pulse sequence. (B and C) Relative levels of Lac (B) and TGs (C) over time obtained for MDA-MB-435 cells in DMECP or RPMICP. Values presented as mean ± SE are from global 1D 1H MR spectra acquired with a spin echo-based pulse sequence and Lac editing.
Intracellular LacTG levels increased during the time course of the MBC experiments independent of the perfusion medium (Figures 5, B–F, and 6A). For untreated MDA-MB-435 cells in DMECP, LacTG levels increased only until day 1 and leveled out, which was also reflected in the localized 1H MR spectra (Figures 5B and 6A). A comparison with global Lac-edited 1H MR spectra suggests that both intracellular Lac and TG were responsible for the increase in intracellular LacTG with time (Figure 6, B and C). Typically, intracellular metabolite levels appeared to be higher in MDA-MB-435 cells maintained in DMECP than in cells in RPMICP (Figures 5 and 6).
Intracellular tCho and tCr along the sample were unaffected by indomethacin treatment, independent of the cell culture and perfusion medium used (data not shown). However, indomethacin treatment significantly increased LacTG in MDA-MB-435 cells maintained in RPMICP compared to control cells, localized within and directly below the region of the sample containing the indomethacin pellets (Figure 5, D–F). This localized LacTG increase as a result of exposure to indomethacin was not reflected in the global 1D 1H MR spectra of MDA-MB-435 cells in RPMICP (Figure 6A). Global Lac-edited 1D 1H MR spectra of MDA-MB-435 cells in RPMICP revealed that, compared to control cells, indomethacin treatment significantly increased intracellular TG (Figure 6C) but did affect intracellular Lac levels only for MDA-MB-435 cells in RPMICP treated with two pellets of indomethacin between days 1 and 2 (Figure 6B).
Global 1D 1H MR spectra of MDA-MB-435 cells perfused with DMECP did not reveal any significant effect of indomethacin treatment on intracellular concentrations of tCho, tCr, LacTG, Lac, or TG compared to control cells (Figure 6). The absence of indomethacin-induced LacTG changes in MDA-MB-435 cells in DMECP was echoed by only a marginal effect on intracellular LacTG levels in localized MR spectra (Figure 5, B and C).
Results from microarray analysis are displayed in Table 1. Several genes were upregulated or downregulated by 48 hours of low-dose exposure of MDA-MB-435 cells to indomethacin. Currently known functions of each of these genes are also listed in this table. Most of these genes are involved in amino acid metabolism, with only one gene connected to fatty acid metabolism, indicating that the action of indomethacin occurs mainly at the protein level rather than through genetic modulation.
Table 1.
Fold Change (Fc) in the Gene Expression of MDA-MB-435 Cells Cultured in RPMIC in Response to Indomethacin Treatment.
| Affymetrix Probe Set ID* | Fc | P | UniGene ID*, Gene Symbol (Gene Title) | Gene Ontology Biologic Process//Molecular Function | Pathway (KEGG)† |
| 230746_s_at | -2.03 | 1.00 | Hs.25590, STC1 (stanniocalcin 1) | Calcium ion homeostasis (tca)//cell surface receptor-linked signal transduction (tca)//cell-cell signaling (tca)//response to nutrients (tca)//hormone activity (tca) | |
| 209183_s_at | -1.85 | 1.00 | Hs.93675, C10orf10 (chromosome 10 open reading frame 10) | ||
| 237631_at | -1.82 | 1.00 | Hs.195400,-(transcribed locus) | ||
| 239870_at | -1.71 | .92 | Hs.135283, SPATS1 (spermatogenesis-associated, serine-rich 1) | ||
| 200648_s_at | -1.64 | .63 | Hs.518525, GLUL (glutamate-ammonia ligase (glutamine synthase)) | Regulation of neurotransmitter levels (not recorded)//glutamine biosynthesis (ifa)//glutamine biosynthesis (not recorded)//nitrogen compound metabolism (ifa)//glutamate-ammonia ligase activity (ifa)//ligase activity (ifa) | Glutamate metabolism, peptidoglycan biosynthesis, nitrogen metabolism, glutamate receptor signaling pathway (Ingenuity) |
| 231894_at | 1.63 | .63 | Hs.531176, SARS (seryl-tRNA synthetase) | Protein biosynthesis (ifa)//protein biosynthesis (tca)//seryl-tRNA aminoacylation (ifa)//tRNA processing (tca)//RNA binding (tca)//serine-tRNA ligase activity (ifa)//serine-tRNA ligase activity (tca)//ATP binding (ifa)//ligase activity (ifa) | Glycine, serine, and threonine metabolism; aminoacyl-tRNA biosynthesis |
| 204285_s_at | 1.64 | .55 | Hs.96, PMAIP1 (phorbol-12-myristate-13-acetate-induced protein 1) | ||
| 212816_s_at | 1.65 | .81 | Hs.533013, CBS (cystathionine β-synthase) | Cysteine biosynthesis from serine (ifa)//metabolism (ifa)//amino acid biosynthesis (ifa)//cysteine biosynthesis through cystathione (ifa)//cystathionine β-synthase activity (tca)//lyase activity (ifa) | Gycine, serine, threonine metabolism; selenoamino acid metabolism |
| 200924_s_at | 1.66 | .56 | Hs.502769, SLC3A2 (solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2) | Carbohydrate metabolism (ifa)//calcium ion transport (non-(tca))//amino acid transport (tca)//cell growth (non-(tca))//catalytic activity (ifa)//α-amylase activity (ifa)//calcium-sodium antiporter activity (tca) | Starch and sucrose metabolism |
| 204286_s_at | 1.67 | .83 | Hs.96, PMAIP1 (phorbol-12-myristate-13-acetate-induced protein 1) | ||
| 202887_s_at | 1.69 | .60 | Hs.523012, DDIT4 (DNA damage-inducible transcript 4) | ||
| 200670_at | 1.70 | .81 | Hs.437638, XBP1 (X-box-binding protein 1) | Transcription (ifa)//regulation of transcription, DNA-dependent (ifa)//immune response (tca)//transcription factor activity (tca) | Endoplasmic reticulum pathway (Ingenuity) |
| 219270_at | 1.72 | .95 | Hs.155569, MGC4504 (hypothetical protein MGC4504) | ||
| 221156_x_at | 1.74 | .97 | Hs.285051, CCPG1 (cell cycle progression 1) | ||
| 218145_at | 1.74 | .89 | Hs.516826, TRIB3 (tribbles homolog 3 (Drosophila)) | Activation of MAPK (inferred from direct assay)//transcription (ifa)//regulation of transcription, DNA-dependent (ifa)//protein amino acid phosphorylation (ifa)//negative regulation of protein kinase activity (ifsss)//apoptosis (ifsss)//transcription corepressor activity (ifsss)//protein kinase activity (ifa)//protein kinase inhibitor activity (ifa)//protein binding (inferred from physical interaction)//protein binding (ifsss)//ATP binding (ifa)//protein kinase binding (inferred from physical interaction)//protein kinase binding (ifsss) | |
| 209146_at | 1.80 | .99 | Hs.105269, SC4MOL (sterol-C4-methyl oxidase-like) | Fatty acid metabolism (tca)//metabolism (ifa)//sterol biosynthesis (ifa)//C4 methylsterol oxidase activity (tca)//oxidoreductase activity (ifa) | Cholesterol biosynthesis (GenMAPP) |
| 205194_at | 1.80 | .99 | Hs.512656, PSPH (phosphoserine phosphatase) | l-serine biosynthesis (ifa)//metabolism (ifa)//amino acid biosynthesis (ifa)//magnesium ion binding (ifa)//catalytic activity (ifa)//phosphoserine phosphatase activity (ifa)//phosphoserine phosphatase activity (tca)//hydrolase activity (ifa) | Glycine, serine, and threonine metabolism |
| 232689_at | 1.81 | 1.00 | Hs.504501, LOC284561 (hypothetical protein LOC284561) | ||
| 217127_at | 1.85 | 1.00 | Hs.19904, CTH (cystathionase (cystathionine γ-lyase)) | Amino acid biosynthesis (ifa)//cysteine biosynthesis (ifa)//cystathionine γ-lyase activity (ifa)//lyase activity (ifa) | Glycine, serine, and threonine metabolism; methionine metabolism; cysteine metabolism; selenoamino acid metabolism; nitrogen metabolism |
| 209921_at | 1.91 | 1.00 | Hs.6682, SLC7A11 (solute carrier family 7 (cationic amino acid transporter, y+ system) member 11) | Protein complex assembly (not recorded)//transport (ifa)//amino acid transport (ifa)//cystine:glutamate antiporter activity (tca)//amino acid permease activity (ifa) | |
| 221577_x_at | 1.91 | 1.00 | Hs.515258, GDF15 (growth differentiation factor 15) | Signal transduction (tca)//TGF-β receptor signaling pathway (tca)//cell-cell signaling (tca)//cytokine activity (tca)//growth factor activity (ifa) | |
| 217678_at | 1.93 | 1.00 | Hs.6682, SLC7A11 (solute carrier family 7 (cationic amino acid transporter, y+ system) member 11) | Protein complex assembly (not recorded)//transport (ifa)//amino acid transport (ifa)//cystine:glutamate antiporter activity (tca)//amino acid permease activity (ifa) | |
| 205475_at | 2.52 | 1.00 | Hs.7122, SCRG1 (scrapie responsive protein 1) | Neurogenesis (tca) | |
| 226181_at | 2.59 | 1.00 | Hs.34851, TUBE1 (tubulin, epsilon 1) | Microtubule-based movement (ifa)//centrosome cycle (tca)//protein polymerization (ifa)//GTPase activity (ifa)//structural constituent of cytoskeleton (tca)//GTP binding (ifa) | |
| 231202_at | 2.62 | 1.00 | Hs.42572, ALDH1L2 (aldehyde dehydrogenase 1 family, member L2) | One-carbon compound metabolism (ifa)//biosynthesis (ifa)//oxidoreductase activity (ifa)//hydroxymethyl, formyl, and related transferase activity (ifa)//cofactor binding (ifa) | |
| 205047_s_at | 2.69 | 1.00 | Hs.489207, ASNS (asparagine synthetase) | Asparagine biosynthesis (ifa)//asparagine biosynthesis (non-(tca))//glutamine metabolism (ifa)//metabolism (ifa)//amino acid biosynthesis (ifa)//asparagine synthase (glutamine-hydrolyzing) activity (inferred from direct assay)//asparagine synthase (glutamine-hydrolyzing) activity (ifa)//ligase activity (ifa) | Alanine and aspartate metabolism; nitrogen metabolism |
The global concentration of indomethacin released into 15 ml of cell culture medium over a period of 24 hours was 4.44 µmol/(l x 24 hours).
Fc < 0 = downregulated genes; Fc > 0 = upregulated genes; P = probability of a gene being differentially expressed; tca = traceable author statement; ifa = inferred from electronic annotation; ifsss = inferred from sequence or structural similarity.
Gene annotations corresponding to the Affymetrix probeset ID are current as of June 2005.
KEGG: www.kegg.com; Ingenuity: www.ingenuity.com; GenMAPP: www.genmapp.org.
Discussion
Indomethacin treatment reduced the invasion of MDA-MB-435 cells into ECM gel, irrespective of whether DME-based or RPMI 1640-based medium was used for cell culture and cell perfusion. These results are consistent with previous observations by Connolly and Rose [37], where treatment of MDA-MB-435 cells with 55.9 µM indomethacin for 72 hours reduced invasion, as measured in a modified Boyden chamber assay. Indomethacin treatment of MDA-MB-435 cells suppressed cancer cell invasion to an even greater extent in the presence of linoleic acid (LA), which stimulated invasion by control cells [37]. Additionally, treatment with 20 µM indomethacin completely inhibited the LA-stimulated MMP-2 activity of MDA-MB-435 breast cancer cells [38]. In a subsequent in vivo study performed by Connolly et al. [39], MDA-MB-435 tumors displayed reduced growth and metastasis to the lung following treatment with indomethacin. In that study, indomethacin was administered in drinking water at a concentration of 10 and 20 µg/ml, leading to approximately 1.3 and 2.5 mg indomethacin/kg body weight per day, respectively.
The observation of reduced invasion in response to indomethacin treatment is not restricted to breast cancer cells but has also been observed in other types of cancer cells and tumor models. Reich and Martin [40] reported that 18 hours of treatment with 10 and 50 µM indomethacin reduced dose dependently the invasion of mouse melanoma cells and a human fibrosarcoma cell line, as measured by a Boyden chamber assay. Dose-dependent reduction in collagenase IV activity was also detected in these studies. In the same study, the treatment of cells with the lipooxygenase inhibitor esculetin induced similar effects, indicating that COX and lipooxygenase pathways of arachidonic acid metabolism play a role in cancer cell invasion.
Indomethacin treatment above 10 µM decreased migration and invasion in C3L5 murine mammary carcinoma cells, and a 24-hour exposure to > 50 µM indomethacin led to loss of viable cells [41]. Additionally, treatment with indomethacin concentrations as low as 5 µM reduced PGE2 expression levels. In these studies, the treatment of C3L5 cells with 50 µM indomethacin did not significantly influence MMP-2 and MMP-9 activities.
Wang et al. [42] reported that glioma cell invasion into Matrigel (ECMatrix, Chemicon International, Temecula, CA) was significantly reduced by high-dose treatment of indomethacin (500 µM), but not at a lower dose of 50 µM. Consistent with changes in invasion, total MMP-2 and active MMP-2 levels in conditioned media were significantly reduced at an indomethacin concentration of 500 µM but not at 50 µM. Cell migration, however, remained unaffected even at doses of up to 500 µM, and doses above 500 mM resulted in cytotoxicity.
Although previous studies have shown a dose-dependent reduction of invasion with indomethacin treatment for doses above 10 µM [40–42], in our study, the invasion of MDA-MB-435 cells exposed to indomethacin treatment was dose-independent for the two doses of ∼ 0.48 and ∼ 0.32 µmol/(l x 24 hours) in the perfusate. One possible explanation may be that cancer cell invasion becomes dose dependent for doses of indomethacin only beyond a threshold dose, which may also depend on cell type. Indomethacin can also act as a mitochondrial uncoupler, which lowers ATP generation and reduces cell proliferation [7]. Cell growth tended to be reduced by indomethacin treatment in our study. Because cell proliferation was factored into the calculation of the invasion index, changes in invasion observed in this study were independent of cell proliferation.
Our data also demonstrate the importance of basal medium in influencing the ability of breast cancer cells to invade and degrade ECM. Even though cell proliferation was comparable for the two media used, MDA-MB-435 cells were significantly more invasive in RPMI than in DME. Studies investigating cancer cell invasion often use serum-containing media, growth factors, or conditioned media as chemoattractants [43]. To date, little is known about the effects of basal medium composition on cancer cell invasion. Haugland and Tysnes [44] used an in vitro coculture assay of tumor spheroids and fetal rat brain cell aggregates as target tissues to assess the invasive ability of three human glioma cell lines cultured in a variety of defined cell culture media and one serum-containing cell culture medium. The study found that the invasive ability of the three cell lines was preserved in all culture media that were able to support spheroid growth, although to varying degrees. Interestingly, directional cell migration on a poly-d-lysine substrate was only observed with the serum-containing culture medium, but not with defined media. A comparison of the composition of DME and RPMI 1640 revealed that the basal medium RPMI 1640 contained l-asparagine (anhydrase), l-aspartic acid, l-glutamic acid, hydroxy-l-proline, d-biotin, p-amino-benzoic acid, and vitamin B12, whereas these amino acids and vitamins were absent in DME. However, most amino acids and vitamins present in both basal media had a higher concentration in DME. In RPMI 1640, Ca(NO3)2·4H2O was the source of calcium and nitrate, whereas DME contained ferric nitrate and calcium chloride, adding iron to inorganic salt components. An additional component missing in DME but present in RPMI 1640 was reduced glutathione, a potent intracellular antioxidant [45]. In contrast, DME contained 2.25-fold more d-glucose than RPMI 1640. It has been reported for a murine fibrosarcoma and a murine squamous cell carcinoma cell line that complete glucose removal from the culture medium increased invasion [46], indicating a role for glucose availability in invasion. However, in our study, both media contained glucose and, based on the levels of the energy phosphates β-NTP and PCr, cellular glucose starvation was not evident in the MR experiments. These data do demonstrate that any interpretation of the invasiveness of cancer cell lines must also take into consideration medium and perfusion conditions. The exact components that caused the increased invasion in cells cultured in RPMI-based medium remain to be identified.
Low-dose indomethacin treatment of MDA-MB-435 cells in DMECP attenuated the drop of GPC levels observed in untreated cells over time and also decreased PC levels of MDA-MB-435 cells in RPMICP. These data are consistent with results obtained from HMEC extracts, intact breast cancer cells, and extracts of rat stomach [15–18,47]. Proton MR spectra obtained from HMEC extracts exhibited increased GPC and decreased PC following treatment with indomethacin [16]. Consistent with the study on cell extracts, a drop in PC and an increase in GPC were evident in intact MDA-MB-231 cells within 80 minutes of exposing the cells to 200 µM indomethacin [17]. Ramadan et al. [47] observed that GPC increased in indomethacin-induced rat stomach ulcers. Thus, increased GPC appears to be directly related to indomethacin treatment.
An increase in GPC has been previously associated with the reduced malignancy of HMECs in vitro and in vivo [13,48]. These findings agree with our observations that decreased invasion of MDA-MB-435 cells was accompanied by higher intracellular GPC levels.
LacTG levels increased in indomethacin-treated MDA-MB-435 cells in RPMICP, but not DMECP, compared to control cells. This increase was due to increased intracellular TG levels, as intracellular Lac levels did not change significantly with indomethacin treatment.
The increase in TGs observed here is consistent with an earlier in vivo study by Comai et al. [49] where TG content and number of lipid droplets increased in renal medullary interstitial cells of indomethacin-treated rabbits. Increased intracellular TGs have been associated with the induction of apoptosis [50] and have been detected in response to differentiation therapy [51]. The observed increase in intracellular TGs during indomethacin treatment may be the result of both the increased availability of free arachidonic acid and the decrease in prostaglandins due to COX inhibition.
It has been observed in several human cancer cell lines [52–56] and endothelial cells [57] that excess arachidonic acid administered exogenously into the culture medium was rapidly incorporated into TGs and, to a lesser extent, into phospholipids, with only small amounts into diacylglycerides. Additionally, as observed in human neuroblastoma cells, the total TG mass per cell increased, whereas total phospholipid content was not significantly affected [53]. These results, together with observations made by Spinedi [53] and Blank et al. [58] on HL-60 cells, led to the hypothesis that excess free arachidonate is incorporated into TGs through the de novo biosynthesis of glycerolipids by acyl-CoA acyltransferases and that TGs are storage sites for excess cellular arachidonate.
Microarray analyses revealed the downregulation of 5 genes and the upregulation of 19 genes following low-dose treatment with indomethacin. Within the constraints of the current study, none of the currently known direct targets of invasion or metabolism was identified, suggesting that changes in invasion-related enzymes did not occur at the transcriptional level, at least for 2D culture. However, several of the genes identified, such as SLC3A2 or GDF15, are indirectly involved in both functions.
Of the five downregulated genes, the gene STC1 has been found to be upregulated in a majority of breast tumor samples [59]. Here, indomethacin treatment reduced STC1 gene expression by ∼ 2-fold. However, Baek and Eling [21] have shown an increased STC1 gene expression with COX inhibition in human colorectal cancer cells. It is possible that differences in dosage or variability between cell lines may be responsible for this contrasting result. The gene GLUL, also downregulated by indomethacin treatment, encodes for glutamine synthase—an enzyme whose upregulated expression has been associated with shorter disease-free survival in patients with hepatocellular carcinoma [60].
Several upregulated genes identified in microarray data have a broad-ranging impact on cellular pathways, such as amino acid metabolism and molecular functions, and may have played a role in the changes in invasion and metabolism observed here.
Of these upregulated genes, CBS, CTH, SARS, and PSPH encode for proteins that impact on serine metabolism. The enzyme encoded by PSPH regulates the conversion between l-serine and O-phospho-l-serine. The protein encoded by SARS converts l-serine to l-seryl-tRNA/ser and has been shown to induce the cell migration of a CC chemokine receptor 3-transfected human embryonic kidney epithelial cell line [61]. The protein encoded by CBS converts l-serine to l-cystathionine or l-cysteine, with the protein encoded by CTH handling the conversion between l-cysteine and l-cystathionine, and vice versa. Additionally, the proteins encoded by CBS and CTH promote conversion from selenohomocysteine to selenocystathionine to selenocysteine. The genes SLC3A2 and SLC7A11 encode for the membrane proteins 4F2hc and xCt, which form a heterodimeric complex exchanging extracellular anionic cystine for glutamate with a stoichiometry of 1:1 sodium independently [62]. It has been suggested that 4F2hc, a protein encoded by SLC3A2, plays a role in integrin function and is involved in multiple pathways related to cell growth, cell adhesion, and malignant transformation [63]. In addition, glutamate synthesis appeared to be affected by indomethacin treatment because the genes ASNS and CTH encoding for proteins involved in glutamate production were upregulated.
The increased expression of ASNS, TRIB3, and XBP1 by indomethacin treatment indicated a response to endoplasmic reticulum stress at the transcriptional level [64–66]. To what extent these changes at the transcriptional level impact on cell proliferation or growth arrest, apoptosis, or antiapoptosis following indomethacin treatment requires further study. Low-dose indomethacin treatment also upregulated two additional genes PMAIP1 and DDIT4, which encode for proteins involved in apoptosis [67,68].
GDF15, also known as GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, or PTGFB, encodes for a protein that is a member of the transforming growth factor (TGF)-β superfamily and is involved in tissue differentiation and maintenance. The GDF15 protein, macrophage inhibitory cytokine 1, has been shown to be elevated in the serum of patients with metastatic prostate, breast, and colorectal carcinomas [69]. The GDF15 protein is involved in apoptosis and other biologic functions in an autocrine or a paracrine manner, independent of COX regulation [21]. Indomethacin treatment increased GDF15 mRNA levels dose dependently and time dependently in human colorectal, breast, prostate, lung epithelial, and oral cavity cancer cells [22,70]. The GDF15 gene is regulated in response to a number of NSAIDs in several cancer cell lines through COX-independent and p53-independent mechanisms [22]. The primary role of GDF15 in cancer cell invasion is yet to be determined.
In our study, treatment of MDA-MB-435 breast cancer cells with 4.44 µmol/(l x 24 hours) for 24 hours did not affect MMP gene expression. It is possible that indomethacin may inhibit MMP expression through repression of gene transcription because treatment of lung cancer cells with 20 and 100 µM indomethacin for 24 hours reduced the promoter activity of MMP-2 to ∼ (80+10)% and ∼ (20+2)% of control, respectively [71]. Treatment of MDA-MB-231 cells with a short-term high dose of 300 µM indomethacin for 2 hours resulted in the underexpression or overexpression of multiple genes involved in processes such as angiogenesis, apoptosis, cell adhesion, cell cycle regulation, cell differentiation, cell motility, choline phospholipid metabolism, and DNA repair [18]. Again, none of the known genes directly involved in choline phospholipid metabolism were identified in this study. Interestingly, there was no overlap in the changes in gene expression profiles observed by Glunde et al. [18] for short-term high-dose treatment of MDA-MB-231 cells and long-term low-dose treatment of MDA-MB-435 cells in the current study, which may be explained, in part, by differences in cell type and dose.
In summary, we have shown here that a nonspecific COX inhibitor, indomethacin, induced a profound reduction in the ability of breast cancer cells to invade and degrade ECM gel. Metabolic changes in choline phospholipids and TGs more typical of differentiated HMECs were temporally coincident with changes in invasion. Microarray data revealed several molecular changes that may have played a role in the reduced invasion and altered metabolism observed here and could potentially represent novel COX-independent target genes in the invasion pathway. Our data strongly support the use of anti-inflammatory agents in reducing breast cancer invasion.
Acknowledgements
We thank V. P. Chacko and J. Gillen for their expert technical assistance. We are indebted to D. C. Shungu and X. Mao for the NMR data processing software XsOsNMR.
Abbreviations
- CHESS
CHEmical Shift Selective
- DMEC
DME plus supplements
- DMECP
DMEC plus 10 mM HEPES
- ΔPi
chemical shift of inorganic phosphate
- ECM
extracellular matrix
- FID
free induction decay
- GPC
glycerophosphocholine
- LA
linoleic acid
- LacTG
lactate + triglyceride
- MR
magnetic resonance
- MBC
Metabolic Boyden Chamber
- PC
phosphocholine
- Pi
inorganic phosphate (ex, extracellular; in, intracellular)
- RPMI
Roswell Park Memorial Institute
- RPMIC
RPMI 1640 plus supplements
- RPMICP
RPMIC plus 10 mM HEPES
- tCho
total choline (choline + phosphocholine + glycerophosphocholine)
- tCr
total creatine (creatine + phosphocreatine)
Footnotes
This work was supported by National Institutes of Health grant RO1 CA82337.
References
- 1.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 2.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartsburd PM. Chronic inflammation as inductor of procancer microenvironment: pathogenesis of dysregulated feedback control. Cancer Metastasis Rev. 2003;22:95–102. doi: 10.1023/a:1022220219975. [DOI] [PubMed] [Google Scholar]
- 4.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashfi K, Rigas B. Non-COX-2 targets and cancer: expanding the molecular target repertoire of chemoprevention. Biochem Pharmacol. 2005;70:969–986. doi: 10.1016/j.bcp.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Simmons DL, Botting RM, Hla T. Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol Rev. 2004;56:387–437. doi: 10.1124/pr.56.3.3. [DOI] [PubMed] [Google Scholar]
- 7.Fosslien E. Biochemistry of cyclooxygenase (COX)-2 inhibitors and molecular pathology of COX-2 in neoplasia. Crit Rev Clin Lab Sci. 2000;37:431–502. doi: 10.1080/10408360091174286. [DOI] [PubMed] [Google Scholar]
- 8.Noguchi M, Rose DP, Earashi M, Miyazaki I. The role of fatty acids and eicosanoid synthesis inhibitors in breast carcinoma. Oncology. 1995;52:265–271. doi: 10.1159/000227471. [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick FA. Cyclooxygenase enzymes: regulation and function. Curr Pharm Des. 2004;10:577–588. doi: 10.2174/1381612043453144. [DOI] [PubMed] [Google Scholar]
- 10.Rigas B, Kashfi K. Cancer prevention: a new era beyond cyclooxygenase-2. J Pharmacol Exp Ther. 2005;314:1–8. doi: 10.1124/jpet.104.080564. [DOI] [PubMed] [Google Scholar]
- 11.Leslie CC. Regulation of arachidonic acid availability for eicosanoid production. Biochem Cell Biol. 2004;82:1–17. doi: 10.1139/o03-080. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser E, Chiba P, Zaky K. Phospholipases in biology and medicine. Clin Biochem. 1990;23:349–370. doi: 10.1016/0009-9120(90)90051-u. [DOI] [PubMed] [Google Scholar]
- 13.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 14.Ackerstaff E, Pflug BR, Nelson JB, Bhujwalla ZM. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Res. 2001;61:3599–3603. [PubMed] [Google Scholar]
- 15.Natarajan K, Mori N, Artemov D, Aboagye EO, Chacko VP, Bhujwalla ZM. Phospholipid profiles of invasive human breast cancer cells are altered towards a less invasive phospholipid profile by the anti-inflammatory agent indomethacin. Adv Enzyme Regul. 2000;40:271–284. doi: 10.1016/s0065-2571(99)00026-6. [DOI] [PubMed] [Google Scholar]
- 16.Natarajan K, Mori N, Artemov D, Bhujwalla ZM. Exposure of human breast cancer cells to the anti-inflammatory agent indomethacin alters choline phospholipid metabolites and Nm23 expression. Neoplasia. 2002;4:409–416. doi: 10.1038/sj.neo.7900252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glunde K, Ackerstaff E, Natarajan K, Artemov D, Bhujwalla ZM. Real-time changes in 1H and 31P NMR spectra of malignant human mammary epithelial cells during treatment with the anti-inflammatory agent indomethacin. Magn Reson Med. 2002;48:819–825. doi: 10.1002/mrm.10295. [DOI] [PubMed] [Google Scholar]
- 18.Glunde K, Jie C, Bhujwalla ZM. Mechanisms of indomethacin-induced alterations in the choline phospholipid metabolism of breast cancer cells. Neoplasia. 2006;8:758–771. doi: 10.1593/neo.06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackerstaff E, Artemov D, Gillies RJ, Bhujwalla ZM. Evaluation of breast cancer cell invasion with the Metabolic Boyden Chamber; 9th Scientific Meeting and Exhibition of the International Society of Magnetic Resonance in Medicine, ISMRM-ESMRB Joint Annual Meeting; Glasgow, UK. 2001. [Google Scholar]
- 20.Pilatus U, Ackerstaff E, Artemov D, Mori N, Gillies RJ, Bhujwalla ZM. Metabolic Boyden Chamber detects significant differences in invasion for metastatic and nonmetastatic prostate cancer cells; 7th Scientific Meeting and Exhibition of the International Society of Magnetic Resonance in Medicine; Philadelphia, PA, USA. 1999. [Google Scholar]
- 21.Baek SJ, Eling TE. Changes in gene expression contribute to cancer prevention by COX inhibitors. Prog Lipid Res. 2006;45:1–16. doi: 10.1016/j.plipres.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Baek SJ, Wilson LC, Lee CH, Eling TE. Dual function of nonsteroidal anti-inflammatory drugs (NSAIDs): inhibition of cyclooxygenase and induction of NSAID-activated gene. J Pharmacol Exp Ther. 2002;301:1126–1131. doi: 10.1124/jpet.301.3.1126. [DOI] [PubMed] [Google Scholar]
- 23.Cailleau R, Olive M, Cruciger QV. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14:911–915. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 24.Sanford KK, Earle WR, Evans VJ, Waltz HK, Shannon JE. The measurement of proliferation in tissue cultures by enumeration of cell nuclei. J Natl Cancer Inst. 1950;11:773–788. [PubMed] [Google Scholar]
- 25.Pilatus U, Ackerstaff E, Artemov D, Mori N, Gillies RJ, Bhujwalla ZM. Imaging prostate cancer invasion with multi-nuclear magnetic resonance methods: the Metabolic Boyden Chamber. Neoplasia. 2000;2:273–279. doi: 10.1038/sj.neo.7900089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilatus U, Aboagye E, Artemov D, Mori N, Ackerstaff E, Bhujwalla ZM. Real-time measurements of cellular oxygen consumption, pH, and energy metabolism using nuclear magnetic resonance spectroscopy. Magn Reson Med. 2001;45:749–755. doi: 10.1002/mrm.1102. [DOI] [PubMed] [Google Scholar]
- 27.Hetherington HP, Hamm JR, Pan JW, Rothman DL, Shulman RG. A fully localized h-1 homonuclear editing sequence to observe lactate in human skeletal-muscle after exercise. J Magn Res. 1989;82:86–96. [Google Scholar]
- 28.Prichard JW, Alger JR, Behar KL, Petroff OA, Shulman RG. Cerebral metabolic studies in vivo by 31P NMR. Proc Natl Acad Sci USA. 1983;80:2748–2751. doi: 10.1073/pnas.80.9.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corbett R, Laptook A, Weatherall P. Noninvasive measurements of human brain temperature using volume-localized proton magnetic resonance spectroscopy. J Cereb Blood Flow Metab. 1997;17:363–369. doi: 10.1097/00004647-199704000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Young IR, Bell JD, Hajnal JV, Jenkinson G, Ling J. Evaluation of the stability of the proton chemical shifts of some metabolites other than water during thermal cycling of normal human muscle tissue. J Magn Reson Imaging. 1998;8:1114–1118. doi: 10.1002/jmri.1880080517. [DOI] [PubMed] [Google Scholar]
- 31.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 32.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J Comput Biol. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- 35.Kendziorski CM, Newton MA, Lan H, Gould MN. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat Med. 2003;22:3899–3914. doi: 10.1002/sim.1548. [DOI] [PubMed] [Google Scholar]
- 36.Newton MA, Kendziorski CM. Parametric empirical Bayes methods for microarrays. In: Parmigiani G, Garrett ES, Irizarry R, Zeger sL, editors. The Analysis of Gene Expression Data: Methods and Software. New York: Springer Verlag; 2003. p. 504. [Google Scholar]
- 37.Connolly JM, Rose DP. Effects of fatty acids on invasion through reconstituted basement membrane (“Matrigel”) by a human breast cancer cell line. Cancer Lett. 1993;75:137–142. doi: 10.1016/0304-3835(93)90198-i. [DOI] [PubMed] [Google Scholar]
- 38.Liu XH, Rose DP. Stimulation of type IV collagenase expression by linoleic acid in a metastatic human breast cancer cell line. Cancer Lett. 1994;76:71–77. doi: 10.1016/0304-3835(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 39.Connolly JM, Liu XH, Rose DP. Dietary linoleic acid-stimulated human breast cancer cell growth and metastasis in nude mice and their suppression by indomethacin, a cyclooxygenase inhibitor. Nutr Cancer. 1996;25:231–240. doi: 10.1080/01635589609514447. [DOI] [PubMed] [Google Scholar]
- 40.Reich R, Martin GR. Identification of arachidonic acid pathways required for the invasive and metastatic activity of malignant tumor cells. Prostaglandins. 1996;51:1–17. doi: 10.1016/0090-6980(95)00154-9. [DOI] [PubMed] [Google Scholar]
- 41.Rozic JG, Chakraborty C, Lala PK. Cyclooxygenase inhibitors retard murine mammary tumor progression by reducing tumor cell migration, invasiveness and angiogenesis. Int J Cancer. 2001;93:497–506. doi: 10.1002/ijc.1376. [DOI] [PubMed] [Google Scholar]
- 42.Wang M, Yoshida D, Liu S, Teramoto A. Inhibition of cell invasion by indomethacin on glioma cell lines: in vitro study. J Neuro-Oncol. 2005;72:1–9. doi: 10.1007/s11060-004-1392-0. [DOI] [PubMed] [Google Scholar]
- 43.Albini A, Benelli R, Noonan DM, Brigati C. The “chemoinvasion assay”: a tool to study tumor and endothelial cell invasion of basement membranes. Int J Dev Biol. 2004;48:563–571. doi: 10.1387/ijdb.041822aa. [DOI] [PubMed] [Google Scholar]
- 44.Haugland HK, Tysnes OB. Heterogenic modulation of malignant behavior in human glioma cells in defined and serum-containing media. In Vitro Cell Dev Biol Anim. 1996;32:159–166. doi: 10.1007/BF02723681. [DOI] [PubMed] [Google Scholar]
- 45.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 46.Cuvier C, Jang A, Hill RP. Exposure to hypoxia, glucose starvation and acidosis: effect on invasive capacity of murine tumor cells and correlation with cathepsin (L + B) secretion. Clin Exp Metastasis. 1997;15:19–25. doi: 10.1023/a:1018428105463. [DOI] [PubMed] [Google Scholar]
- 47.Ramadan S, Bonin AM, Kennedy BJ, Hambley TW, Lay PA. Nuclear magnetic resonance analysis of indomethacin-induced gastric ulcers. Chem Res Toxicol. 2005;18:123–128. doi: 10.1021/tx049806t. [DOI] [PubMed] [Google Scholar]
- 48.Bhujwalla ZM, Aboagye EO, Gillies RJ, Chacko VP, Mendola CE, Backer JM. Nm23-transfected MDA-MB-435 human breast carcinoma cells form tumors with altered phospholipid metabolism and pH: a 31P nuclear magnetic resonance study in vivo and in vitro. Magn Reson Med. 1999;41:897–903. doi: 10.1002/(sici)1522-2594(199905)41:5<897::aid-mrm7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 49.Comai K, Prose P, Farber SJ, Paulsrud JR. Correlation of renal medullary prostaglandin content and renal interstitial cell lipid droplets. Prostaglandins. 1974;6:375–379. doi: 10.1016/0090-6980(74)90010-0. [DOI] [PubMed] [Google Scholar]
- 50.Hakumaki JM, Kauppinen RA. 1H NMR visible lipids in the life and death of cells. Trends Biochem Sci. 2000;25:357–362. doi: 10.1016/s0968-0004(00)01614-5. [DOI] [PubMed] [Google Scholar]
- 51.Milkevitch M, Shim H, Pilatus U, Pickup S, Wehrle JP, Samid D, Poptani H, Glickson JD, Delikatny EJ. Increases in NMR-visible lipid and glycerophosphocholine during phenylbutyrate-induced apoptosis in human prostate cancer cells. Biochim Biophys Acta. 2005;1734:1–12. doi: 10.1016/j.bbalip.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Desplat V, Dulery C, Praloran V, Denizot Y. Incorporation and effect of arachidonic acid on the growth of human myeloma cell lines. Mediat Inflamm. 1999;8:115–118. doi: 10.1080/09629359990612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spinedi A. Different contribution of phospholipid and triacylglycerol metabolism to esterification of free intracellular arachidonate: a study on SK-N-BE(2) human neuroblastoma cells. J Neuro-Oncol. 1997;31:133–139. doi: 10.1023/a:1005714320865. [DOI] [PubMed] [Google Scholar]
- 54.Balsinde J, Dennis EA. The incorporation of arachidonic acid into triacylglycerol in P388D1 macrophage-like cells. Eur J Biochem. 1996;235:480–485. doi: 10.1111/j.1432-1033.1996.00480.x. [DOI] [PubMed] [Google Scholar]
- 55.Butcher RD, Wojcik SJ, Lints T, Wilson T, Schofield PC, Ralph R. Arachidonic acid, a growth signal in murine P815 mastocytoma cells. Cancer Res. 1993;53:3405–3410. [PubMed] [Google Scholar]
- 56.Denizot Y, Najid A, Rigaud M. Incorporation of arachidonic acid in a human cancer gastric tumor cell line (HGT) at various stages of cell proliferation. Cancer Lett. 1993;68:199–205. doi: 10.1016/0304-3835(93)90147-2. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn H, Ponicke K, Halle W, Wiesner R, Schewe T, Forster W. Metabolism of [1–14C]-arachidonic acid by cultured calf aortic endothelial cells: evidence for the presence of a lipoxygenase pathway. Prostaglandins Leukot Med. 1985;17:291–303. doi: 10.1016/0262-1746(85)90118-0. [DOI] [PubMed] [Google Scholar]
- 58.Blank ML, Smith ZL, Snyder F. Arachidonate-containing triacylglycerols: biosynthesis and a lipolytic mechanism for the release and transfer of arachidonate to phospholipids in HL-60 cells. Biochim Biophys Acta. 1993;1170:275–282. doi: 10.1016/0005-2760(93)90010-7. [DOI] [PubMed] [Google Scholar]
- 59.McCudden CR, Majewski A, Chakrabarti S, Wagner GF. Colocalization of stanniocalcin-1 ligand and receptor in human breast carcinomas. Mol Cell Endocrinol. 2004;213:167–172. doi: 10.1016/j.mce.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 60.Osada T, Nagashima I, Tsuno NH, Kitayama J, Nagawa H. Prognostic significance of glutamine synthetase expression in unifocal advanced hepatocellular carcinoma. J Hepatol. 2000;33:247–253. doi: 10.1016/s0168-8278(00)80365-7. [DOI] [PubMed] [Google Scholar]
- 61.Howard OM, Dong HF, Yang D, Raben N, Nagaraju K, Rosen A, Casciola-Rosen L, Hartlein M, Kron M, Yiadom K, et al. Histidyl-tRNA synthetase and asparaginyl-tRNA synthetase, autoantigens in myositis, activate chemokine receptors on T lymphocytes and immature dendritic cells. J Exp Med. 2002;196:781–791. doi: 10.1084/jem.20020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verrey F, Closs EI, Wagner CA, Palacin M, Endou H, Kanai Y. CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. 2004;447:532–542. doi: 10.1007/s00424-003-1086-z. [DOI] [PubMed] [Google Scholar]
- 63.Palacin M, Kanai Y. The ancillary proteins of HATs: SLC3 family of amino acid transporters. Pflugers Arch. 2004;447:490–494. doi: 10.1007/s00424-003-1062-7. [DOI] [PubMed] [Google Scholar]
- 64.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4:966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 66.Kilberg MS, Barbosa-Tessmann IP. Genomic sequences necessary for transcriptional activation by amino acid deprivation of mammalian cells. J Nutr. 2002;132:1801–1804. doi: 10.1093/jn/132.7.1801. [DOI] [PubMed] [Google Scholar]
- 67.Yakovlev AG, Di Giovanni S, Wang G, Liu W, Stoica B, Faden AI. BOK and NOXA are essential mediators of p53-dependent apoptosis. J Biol Chem. 2004;279:28367–28374. doi: 10.1074/jbc.M313526200. [DOI] [PubMed] [Google Scholar]
- 68.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welsh JB, Sapinoso LM, Kern SG, Brown DA, Liu T, Bauskin AR, Ward RL, Hawkins NJ, Quinn DI, Russell PJ, et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc Natl Acad Sci USA. 2003;100:3410–3415. doi: 10.1073/pnas.0530278100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim KS, Yoon JH, Kim JK, Baek SJ, Eling TE, Lee WJ, Ryu JH, Lee JG, Lee JH, Yoo JB. Cyclooxygenase inhibitors induce apoptosis in oral cavity cancer cells by increased expression of nonsteroidal anti-inflammatory drug-activated gene. Biochem Biophys Res Commun. 2004;325:1298–1303. doi: 10.1016/j.bbrc.2004.10.176. [DOI] [PubMed] [Google Scholar]
- 71.Pan MR, Chuang LY, Hung WC. Non-steroidal antiinflammatory drugs inhibit matrix metalloproteinase-2 expression via repression of transcription in lung cancer cells. FEBS Lett. 2001;508:365–368. doi: 10.1016/s0014-5793(01)03118-0. [DOI] [PubMed] [Google Scholar]