Abstract
INSM1 is a downstream target gene of ngn3. A promoter construct containing the −426/+40bp region transiently co-transfected into NIH-3T3 cells with a ngn3 expression plasmid resulted in a 12 fold increase in promoter activity. The ngn3/E47 heterodimer selectively binds and activates the E-box3 of the INSM1 promoter. The endogenous ngn3 and CBP co-activator occupy the INSM1 promoter, resulting in hyper-acetylation of histone H3/H4 chromatin in a human neuroblastoma cell line, IMR-32. Additionally, adenoviral ngn3 can induce endogenous INSM-1 expression in PANC-1 cells through the recruitment of CBP to the INSM1 promoter and increase the acetylation of the INSM1 promoter region.
Keywords: Neurogenin 3, INSM1, CBP, Histone acetylation, Transcriptional activation
1. Introduction
Both the exocrine and endocrine pancreas develops from a common multi-potent progenitor cell (1). During the specification of the pancreatic endocrine cells, a complex cascade of transcription factors must be coordinated to allow proper development of the four distinct hormone-producing cells (reviewed in (2;3) . A key factor in endocrine pancreas specification is the basic helix-loop-helix factor (bHLH) neurogenin 3 (ngn3). Ngn3 expression is both necessary and sufficient to program undifferentiated progenitor cells to an endocrine cell fate (4–6).
Recently the transcriptional repressor, INSM1, has been implicated in the cascade of transcription factors essential in endocrine pancreas development (7). INSM1 was shown to be a direct target of ngn3 regulation and required for the differentiation of the insulin and glucagon producing cells of the endocrine pancreas (8). INSM1 was originally identified from an insulinoma subtraction library (9). Initial expression analysis revealed expression of INSM1 in tumors of neuroendocrine origin (9;10). Additionally, INSM1 expression was observed in early fetal pancreas (7;8;11) and fetal brain (12;13) development. Transgenic mouse studies using a ~1.7 kb region of the INSM1 promoter demonstrated early restricted expression of the INSM1 gene as early as e9.5 in the developing CNS, olfactory bulb, olfactory neuroepithelium, trigerminal ganglia, retina neuron, and spinal cord (12). We identified NeuroD/β2 as a regulatory protein of the INSM1 promoter (12). NeuroD/β2 is expressed during early pancreas and brain development. NeuroD/β2 is activated by ngn3 and persists in the mature islet cells where it plays a role in maintaining the differentiated state (14) and regulates the expression of insulin (15) and glucagon (16).
In this study, we investigate the molecular mechanisms underlying the induction of the INSM1 gene by ngn3. Transient transfections and EMSA analyses demonstrated specific binding of the ngn3/E47 heterodimer on the E-box3 of the INSM1 promoter. Using chromatin immunoprecipitation (ChIP) assay, we showed binding of endogenous ngn3 protein on the INSM1 promoter region. Further, over-expression of ngn3 protein induced endogenous INSM1 expression in a pancreatic ductal carcinoma cell line, PANC-1. Using additional ChIP assays, we could conclusively demonstrate that the CBP co-activator was recruited to the same INSM1 promoter sequence with a coordinate increase in acetylation both in IMR-32 (endogenous ngn3) and in Ad-ngn3 infected PANC-1 cells.
2. Materials and Methods
2.1 Cell culture and transient transfection
NIH-3T3, IMR-32, S-KN-MC, U118MG, U87MG, D283Med, Daoy, and PANC-1 were maintained in the designated medium as recommended (ATCC). For transient transfection studies, the cells were transfected with lipofectamine 2000 reagent (Life Technologies) as described (12). Forty-eight hours post-transfection the cells were collected, lysed and assayed for CAT (Promega) activities and normalized with β-galactosidase. All experiments were repeated at least three times and the average with the SEM are shown.
2.2 DNA constructs
The –426/+40 bp construct was created by NheI digestion of the –1661/+40 bp INSM1p-CAT3 construct. The INSM1pΔE-box3 CAT construct was prepared as previously described (12). The E47 expression construct was a kind gift from Dr. Roland Stein (Vanderbilt University). The pCR3.1β2 (NeuroD/β2 expression vector) construct was kindly provided by Dr. M. J Tsai (Baylor College of Medicine). The pcDNA3-ngn3 construct was generated by PCR amplification from mouse fetal pancreas with sense primer 5’-GTAAGCTTCCAACCGCAGGATG GCGCCTCATCCTT-3’ and anti-sense primer 5’-GCGGATCCGTCTCTTCACAAGAAGTCTGAGAACACC-3’ containing a HindIII or BamHI site (bold) in each primer to enable cloning into the pcDNA3 expression plasmid (Invitrogen). The mouse ngn3 cDNA was confirmed by sequencing. The 3X E-box constructs were created using synthetic oligonucleotides containing three tandem copies of the individual E-box elements as described (12).
2.3 Electrophoretic Mobility Shift Assay (EMSA)
EMSA analysis was performed using a double strand oligonucleotide spanning the –192 to –165 bp region of the INSM1 promoter (containing E-box3), 5’ CCCTCAGGTACATCTGCCGCACCTACCG 3” and the complementary strand as described (12). The double strand oligonucleotide was end-labeled using γ-32P-ATP (3000 Ci/mmol, NEN) and T4 polynucleotide kinase (New England Biolabs). E47, NeuroD/β2, and ngn3 proteins were synthesized using the expression plasmids pcDNA3-ngn3, pCR3.1-NeuroD/β2, and pcDNA3-E47 with the TNT-coupled rabbit reticulocyte lysate kit (Promega). Supershift experiments were performed using 1μg each of mouse anti-ngn3 antibody (Developmental Studies Hybridoma Bank, University of Iowa), rabbit anti-NeuroD/β2 antibody (Santa Cruz Biotech.), or control species matched IgG (Southern Biotechnologies). For competition experiments, 50 or 100 molar excess of cold 3X-E1, 3X-E2, or 3X-E3 oligonucleotide was added along with the labeled oligonucleotide. The protein-DNA complexes were resolved on a 4% PAGE (40:1) gel. The gels were dried and exposed to autoradiography.
2.4 RT-PCR
Total RNA was isolated from human tumor cells lines, IMR-32, SK-N-MC (neuroblastoma), U87MG, U118MG (glioblastoma), D283Med, Daoy (medulloblastoma), and PANC-1 (pancreatic carcinoma) using Trizol reagent according to manufacturer’s protocol (Invitrogen). PANC-1 cells were transduced with either Ad-LacZ or Ad-ngn3 at an m.o.i. of 100:1 for three days. First-strand cDNA was prepared from 5 μg of total RNA with the SuperScipt II RT kit (Invitrogen). One-tenth of the cDNA was used as a template for a PCR reaction. Human INSM1 primers are (Forward) 5’-AACTGTCCTTCGCTTGGA-3’, (Reverse) 5’-ACGAGACAAACGCGTACAGCT-3’ (80 bp); human GAPDH primers are (Forward) 5’-ACCACAGTCCATGCCATCAC-3’, (Reverse) 5’-TCCACCACCCTGTTGCTGTA-3’ (127 bp); mouse ngn3 primers are (Forward) 5’-TTCGCCCACAACTACATCTG-3’, (Reverse) 5’-CCAGGGAATTCCACCAATGA-3’ (200 bp); human ngn3 primers are (forward) 5’ GGTAGAAAGGATGACGCCTC 3’ and (reverse) 5’ CCGAGTTGAGGTCGTGCAT 3’ (309 bp).
2.5 Construction of recombinant adenovirus
Recombinant adenoviruses were generated using the AdEasy XL adenoviral vector system (Stratagene). Briefly, ngn3 cDNA was subcloned into the pCMV-Shuttle vector as described (17). To determine the adenovirus titer, the BD adeno X rapid titer kit was used to stain the viral infected cells with anti-hexon antibody (BD Biosciences Clontech).
2.6 Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using either human neuroblastoma, IMR-32 or adenovirus transduced PANC-1 cells. A ChIP assay kit (Upstate) was used. The procedures were followed as described previously (17). The pre-cleared chromatin was incubated with anti-ngn3 antibody (Developmental Studies Hybridoma Bank, University of Iowa), anti-CBP (Santa Cruz Biotech.), anti-acetyl-H3, anti-acetyl-H4 (Upstate) or normal mouse/rabbit IgG (Southern Biotechnology Associates, Inc) as negative control. The PCR primers used to detect target sequences were as follows: human INSM1 (Forward) 5’-ATAGAGAAGCAGCAGACCGT-3’, (Reverse) 5’-ATTGTTCTCGCCTCCCGCTT-3’. An additional 5’ upstream INSM1 promoter primer set was used as a negative control (forward) 5’ GAGCTCAAAAGAGCAAGGAA 3’ and (reverse) 5’ AGTCATAAGTTGTGATGGGG 3’. A small amount of input DNA (~2.5%) was used as positive control. Each ChIP assay was performed at least twice to ensure reproducibility.
3. Results and Discussion
3.1 Ngn3 activates E-box3 element of the INSM1 promoter
INSM1 is a transcriptional repressor protein whose expression is limited to early fetal nervous system and fetal pancreas development (7;8;11–13). INSM1 expression can be reactivated in tumors of neuroendocrine origin including insulinoma, pheochromacytoma, retinoblastoma, medulloblastoma, neuroblastoma, medullary thyroid carcinoma, and small cell lung cancer (9;10). Tissue-specific activity of the INSM1 promoter was demonstrated using a transgenic mouse model with the 1.7 kb human INSM1 upstream region driving expression of the LacZ gene (12). Sequential deletion of the promoter region in vitro revealed the importance of a ~500 bp region (−426/+40 bp) for maximal promoter activity (18). Within this region, three E-box elements were identified and E-box3 was shown to be responsible for approximately 60% of the promoter activity that was bound and activated by the NeuroD/β2 -E47 heterodimer (12). NeuroD/β2 has a largely overlapping expression pattern with INSM1 during fetal development as well as in tumors of neuroendocrine origin (12;19). It was speculated that NeuroD/β2 first activated INSM1 while INSM1 could counter-regulate NeuroD/β2 during prenatal pancreas development. Recently, it was demonstrated that over-expression of adenoviral ngn3 in normal adult duct cells induced the expression of INSM1 (8). Therefore, it seems logical to postulate that additional factors must regulate expression of INSM1. Ngn3 expression precedes expression of NeuroD/β2 in the developing endocrine pancreas and is a direct regulator of the NeuroD/β2 gene (19). The sequence of the E-box3 element is also a potential binding site for the pro-endocrine transcription factor ngn3. Therefore, we sought to further identify and define the specific INSM1 promoter region responsible for the induction by ngn3. Transient co-transfection of a −426/+40 bp human INSM1 promoter-CAT construct into NIH-3T3 cells with both ngn3 and E47 expression plasmids caused a ~12 fold increase in the INSM1 promoter-linked reporter activity (Figure 1A). The high level of activation may result from the co-expression of the heterodimer partner, E47. However, addition of E47 alone only resulted in a modest ~2 fold increase in INSM1 promoter activity (Figure 1A). Within this region of the INSM1 promoter there are three potential E-box elements we had previously designated E-box1, E-box2, and E-box3 from most distal to proximal. Using chimeric constructs with three tandem copy repeats of the E-box1, E-box2 or E-box3 upstream of the minimal E1bTATA promoter driven CAT reporter, we sought to demonstrate the E-box(es) responsible for the ngn3 induction. Co-transfection of the 3X E-box1-E1bTATA CAT construct with E47 alone or E47/ngn3 combination showed the same level of induction (~25–30 fold) indicating that E47 could bind and regulate this site (Figure 1B). Co-transfection of the 3X E-box2-E1bTATA CAT construct with E47 alone, ngn3 alone, or E47/ngn3 showed no effect to overall promoter activity (Figure 1B). However, transient transfection of the E47/ngn3 heterodimer with the 3X E-box3-E1bTATA CAT construct showed an approximately 120 fold increase in promoter activity indicating that ngn3/E47 could bind and activate the INSM1 promoter through the E-box3. Additionally, deletion of the E-box3 element from the 500bp INSM1 promoter construct results in an approximately 70% decrease in promoter activity (Figure 1C). Further, co-transfection of the E-box3 deletion construct with E47/ngn3 expression plasmids shows the loss of activation to the INSM1 promoter demonstrating the specificity of the E47/ngn3 complex for this binding site (Figure 1C). Therefore, we have demonstrated that ngn3 and NeuroD/β2 bind to the same site in the INSM1 promoter region. Transient transfections demonstrated that ngn3 is a more potent activator (12 fold verse 5–6 fold) than NeuroD/β2 of INSM1 promoter activity. Both ngn3 and INSM1 expression is transient during early fetal development. INSM1 contains a putative INSM1 binding site and can repress its own expression (20). Although NeuroD/β2 can bind and activate the INSM1 promoter, it is not as potent a regulator as Ngn3. Therefore, in the absence of ngn3, NeuroD/β2 must compete with the negative regulation exerted by INSM1. Therefore, INSM1 likely overcomes the induction by NeuroD/β2 and silences its own expression at later stages of differentiation.
Figure 1.
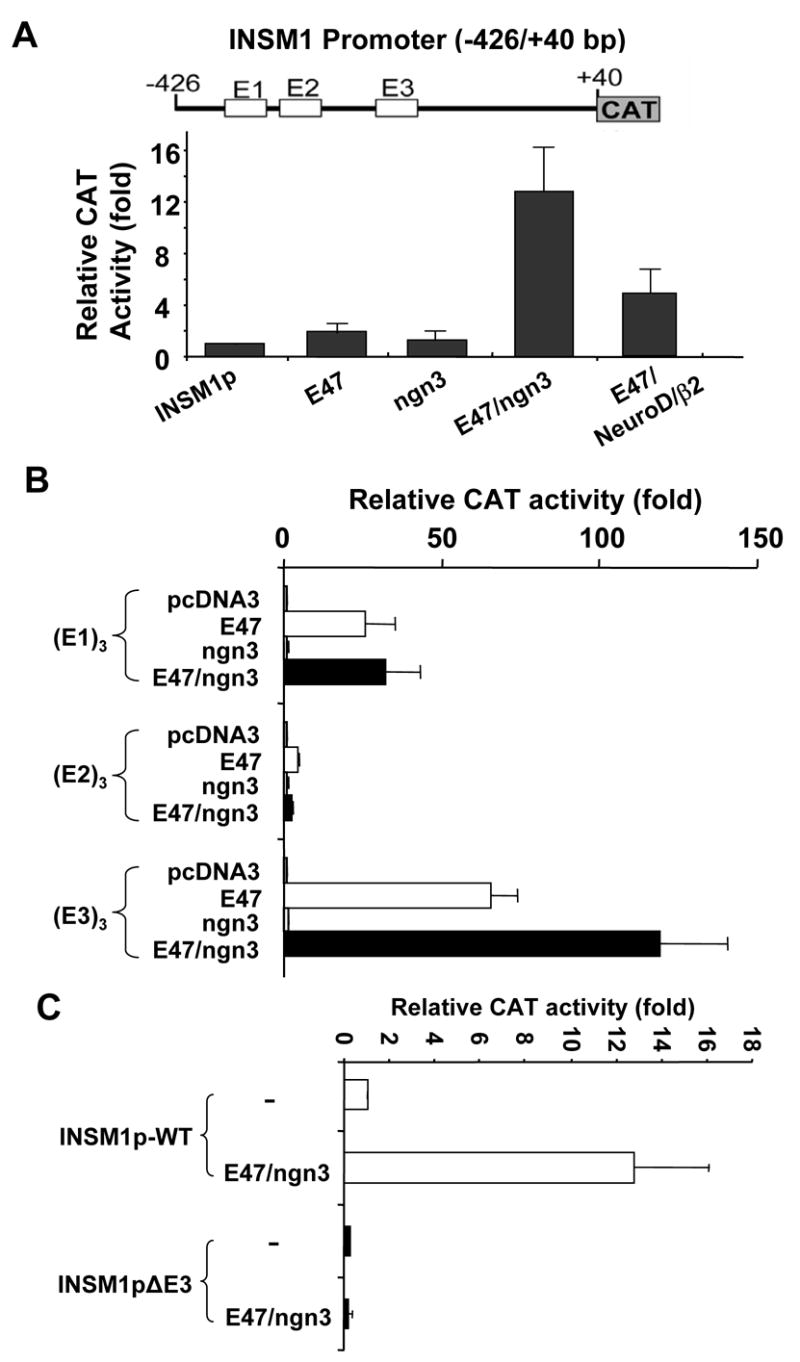
Activation of the INSM1 promoter-CAT constructs. (A) Shown is a schematic diagram of the -426/+40 bp INSM1 promoter CAT construct. The three potential E-box elements are shown as open boxes and indicated as E1, E2, and E3 from most distal to proximal. NIH-3T3 cells were transfected with the -426/+40 bp INSM1 promoter-CAT reporter construct along with either empty expression vector (pcDNA3) or equal molar concentrations of E47, ngn3, and/or NeuroD/β2 expression vectors. Addition of ngn3 and E47 resulted in a 12 fold increase in overall promoter activity. (B) Chimeric constructs containing 3 tandem copy repeats of E1, E2, or E3 box from the INSM1 promoter region were linked to the E1BTATA minimal CAT reporter constructs. Co-transfection of the individual E-box constructs with empty expression vector, E47 alone, ngn3 alone, or E47/ngn3 was performed using NIH-3T3 cells. Shown is the relative fold increase to CAT activity as compared to control. (C) Comparison of the relative activity of the wild type promoter and mutant Ebox3 promoter (INSM1pΔE3) construct with E47/ngn3 expression plasmids were performed in NIH3T3 cells. Deletion of the E-box3 results in a 70% decrease in INSM1 promoter activity. Co-expression of E47/ngn3 with the E-box3 deletion promoter construct shows a complete loss of induction to the promoter activity. Each transfection is repeated at least three times with the calculated SEM shown. All transfections were normalized with the internal control β-galactosidase activity.
3.2 Ngn3/E47 heterodimer physically binds to the E-box3 of the INSM1 promoter
In order to confirm the physical association between ngn3 and the E-box3 of the INSM1 promoter region, we performed an EMSA. E47 alone could bind the E-box3 oligonucleotide (Figure 2A, lane 3), ngn3 alone does not bind (Figure 2A, lane 4) and the addition of equal amounts of E47 and ngn3 formed a heterodimer complex on the E-box3 element seen as a faster migrating complex as compared to the E47 homodimer alone (Figure 2A, lane 5 and lane 3). Additionally, E47 and NeuroD/β2 proteins also form a complex on the same E box element (Figure 2A lane 7 and 9). Supershift experiments demonstrate the specificity of the E47/ngn3 interaction with the E-box3 element. Inclusion of an anti-ngn3 antibody results in a “supershift” in the migration pattern of the E47/ngn3 complex (Figure 2B lane 3). On the contrary, NeuroD/β2 antibody can only partially block NeuroD/β2/E47 complex (Figure 2A lane 10). In order to demonstrate the relative affinities ngn3 and NeuroD/β2 have for the E-box3, we performed an EMSA in the presence of both ngn3 and NeuroD/β2 proteins to determine if there was a preference for binding of one factor. Equal molar amounts of E47, ngn3, and NeuroD/β2 protein were incubated with the E-box3 element and addition of either an anti-ngn3 or an anti-NeuroD/β2 antibody was used to determine which protein formed a complex with the E-box3. Addition of an anti-NeuroD/β2 antibody does not alter the migration of the complex (Figure 2B, lane 7). However, inclusion of an anti-ngn3 antibody shows a supershift complex indicating that ngn3 preferentially binds the E-box3 in the presence of equal molar amounts of NeuroD/β2 protein (Figure 2B, lane 6). Competition experiments were performed to demonstrate the specificity of the interaction between ngn3 and the E-box3 element. Addition of either 50 or 100 molar excess cold competitor 3X E-box3 oligonucleotide completely abolishes binding of the ngn3/E47 protein complex (Figure 2C). Inclusion of 50 or 100 molar excess of 3X E-box1 or 3X E-box2 cold competitor oligonucleotides does not diminish the ngn3/E47-3XE-box3 complex demonstrating that the ngn3 protein interaction with E-box3 is specific.
Figure 2.
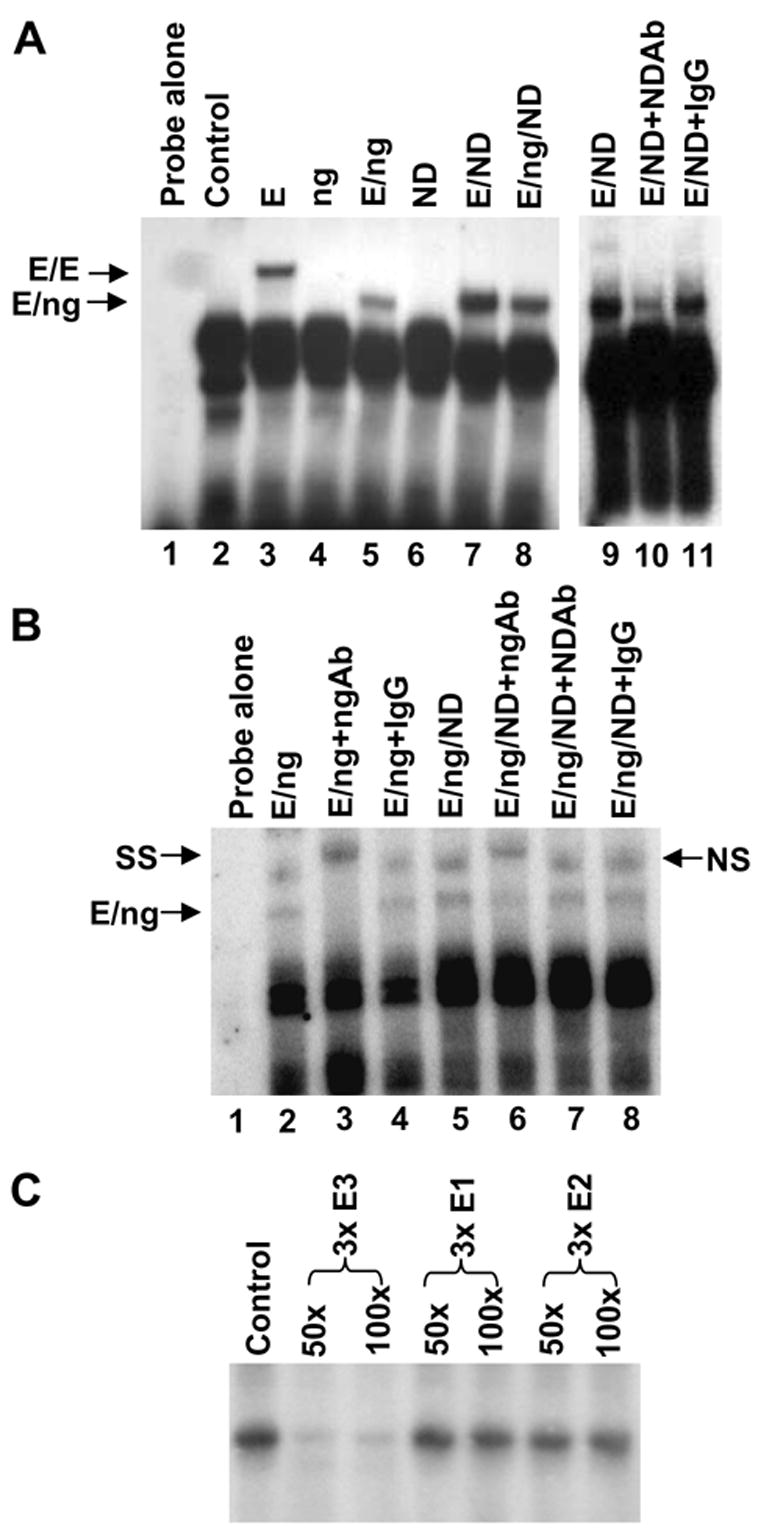
Ngn3 binds to the E-box3. EMSA were performed using in vitro translated protein and an oligonucleotide spanning the −192 to −165bp region derived from the INSM1 promoter. (A) Addition of E47 alone (E), ngn3 (ng) alone or E47/ngn3 (E/ng) proteins results in an E47 homodimer complex and an E47/ngn3 heterodimer complex. E47/NeuroD/β2 proteins (E/ND) form a similar migrating complex. Addition of NeuroD/β2 antibody partially blocks the E/ND complex (lane 10). (B) Supershift experiments were performed in the presence of E47, ngn3, and NeuroD/β2 proteins to determine the relative affinities of each for the E-box3 element. Addition of E47/ngn3 (E/ng) results in a specific complex that is supershifted with the addition of an anti-ngn3 antibody (SS). TNT lysate results in non-specific band as designated NS. Inclusion of an anti-NeuroD/β2 antibody does not alter the complex (lane 7) demonstrating the preference of ngn3 for the E-box3 element. (C) Competition experiments were performed using 32P-labeled −192 to −165bp oligonucleotide bound to E47/ngn3. Fifty or 100 molar excess cold competitor 3X E-box3, 3X E-box1, and 3X E-box2 oligonucleotides were included to demonstrate the specificity of the binding between E-box3 and the E47/ngn3 heterodimer.
3.3 Ngn3 occupies and activates the INSM1 promoter through the recruitment of CBP and hyper-acetylation of histone H3/H4 in IMR-32 cells
We sought to determine if ngn3 was co-expressed with INSM1 in selected neuroendocrine tumor cell lines. We used a small panel of brain tumor cell lines including neuroblastoma IMR-32 (INSM1 positive), SK-N-MC (INSM1 negative), glioblastoma, U87MG and U118MG (INSM1 negative), medulloblastoma, D283Med (INSM1 positive) and Daoy (INSM1 negative) for RT-PCR analysis. Ngn3 is expressed in IMR-32 cells that also co-express INSM1 (Figure 3A, lane 1). However, we did not detect ngn3 in D283Med, another INSM1 positive cell line (Figure 3A, lane 5).
Figure 3.
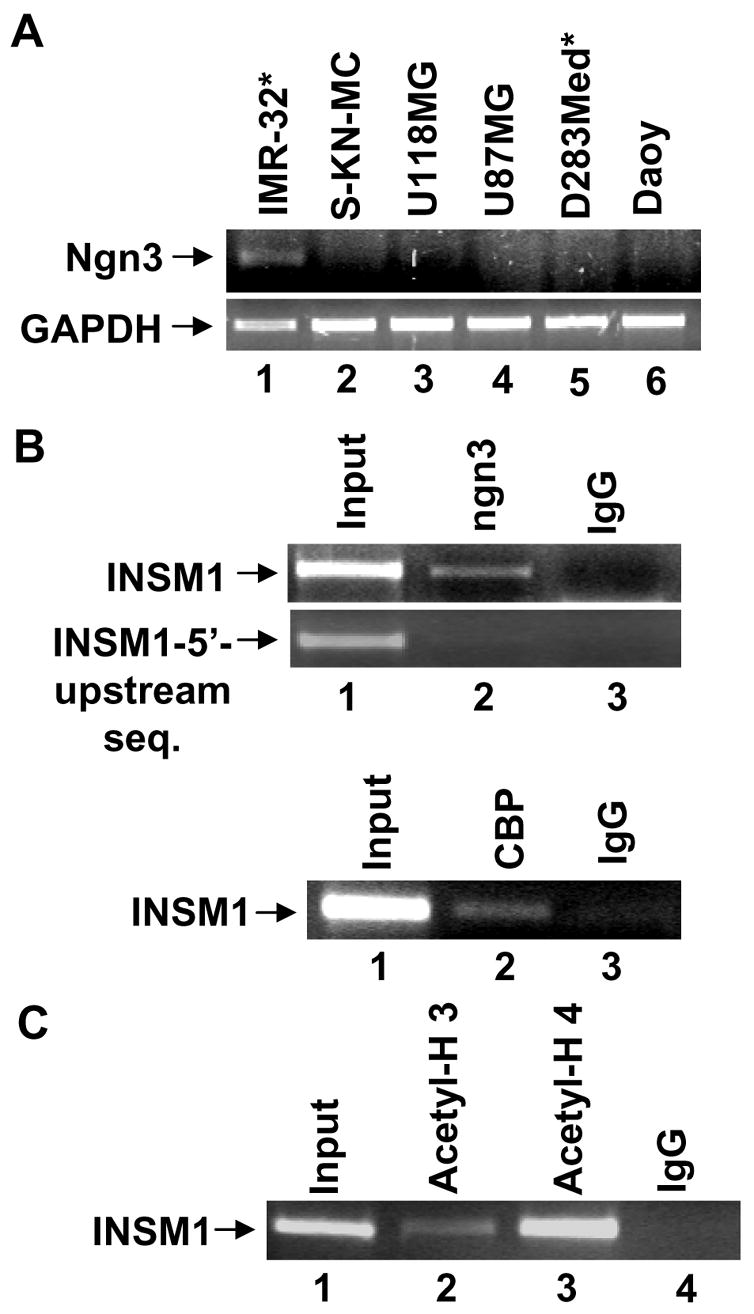
RT-PCR analysis of ngn3, GAPDH, and the ChIP analyses of the endogenous INSM1 promoter. (A) Total RNA was isolated from human neuroblastoma, IMR-32, SK-N-MC, human glioblastoma cells U118MG, U87MG, human medulloblastoma, D283Med, and Daoy using Trizol reagent. Cell lines labeled with * represent the INSM1 expressing cells. IMR-32 cells are positive for ngn3 message. (B) ChIP analyses were performed using a mouse monoclonal anti-ngn3, anti-CBP, anti-acetyl H3/H4, or control IgG antibody with chromatin isolated from IMR-32 cells. Input DNA was included as a positive control (~2.5%) for PCR analysis. The INSM1 promoter region was immunoprecipitated with anti-ngn3, anti-CBP, and anti-acetyl-H3/H4 antibodies but not negative control IgG. Each ChIP experiment was repeated to ensure its reproducibility.
To definitively demonstrate that ngn3 regulates the INSM1 gene via the E-box3 in the -426/+40 bp promoter region, we performed a ChIP assay using a mouse ngn3 monoclonal antibody in IMR-32 cells (both ngn3/INSM1 positive). In the presence of an ngn3 antibody, the region of the INSM1 promoter containing the E-box3 was amplified (Figure 3B, lane 2). The same region of the INSM1 promoter was not amplified with pre-immune mouse IgG (Figure 3B, lane 3). Inclusion of a 5’ upstream INSM1 region that did not contain the E-box3 element was not amplified further demonstrating the specificity of the interaction. Therefore, we showed that endogenous ngn3 protein occupies the E-box3 containing region of the INSM1 promoter in vivo. The recruitment of co-activators such as p300/CBP is required by the bHLH proteins to function as transcriptional activators (21;22). ChIP assays using an anti-CBP antibody demonstrates the recruitment of CBP to the INSM1 promoter region (Figure 3B). The co-activator p300/CBP mediates interactions between the DNA binding transcription factors and the RNA polymerase II transcriptional machinery to facilitate gene transcription (23). Both p300/CBP possess intrinsic acetyltransferase activity and can associate with proteins that possess acetyltransferase activity. Acetylation of histones and other proteins contributes to transcriptional activation and nucleosomal remodeling that accompanies gene activation. In IMR-32 cells, we observed the recruitment of CBP to the INSM1 promoter using anti-CBP antibody. This observation to our knowledge is the first demonstration of ngn3 recruitment of CBP on a bona fide ngn3 target gene. However, other laboratories have demonstrated the direct interaction between ngn3 and p300 when using an artificial E-box construct (24). Furthermore, we have demonstrated that both histone H3 and H4 located at the ngn3 binding region were hyper-acetylated in IMR-32 cells (Figure 3C). Accordingly, these studies enhance our understanding of the modulation of INSM1 by ngn3.
3.4 Ngn3 induces endogenous INSM1 expression in PANC-1 cells through the recruitment of CBP and hyper-acetylation of histone H3/H4
During organ specification, members of the bHLH transcription factor family are required for organogenesis. In pancreas development, a subset of ngn3 positive epithelial cells commit to the endocrine pathway. Our studies are aimed at analyzing the molecular mechanisms for induction of INSM1 by ngn3. In order to confirm that ngn3 can regulate INSM1 in vivo, we used adenoviral expression of ngn3 to induce endogenous expression of INSM1 in human pancreatic ductal epithelial carcinoma cells, PANC-1. RT-PCR analysis with non-transduced, Ad-LacZ, or Ad-ngn3 transduced cells clearly show the induction of INSM1 expression (Figure 4A, lane 3). Ad-ngn3 expression was also confirmed in the PANC-1 cells (Figure 4A, lane 3). ChIP assay using anti-CBP antibody demonstrates the recruitment of CBP to the INSM1 promoter region in the Ad-ngn3 transduced PANC-1 cells in contrast to the Ad-LacZ transduced PANC-1 cells (Figure 4B). Further, ChIP assay was performed to assess the histone acetylation status of the INSM1 promoter region in the presence of over-expressed ngn3. Addition of an anti-acetyl-histone H4 antibody and to a lesser extent anti-acetyl-histone H3 antibody was able to pull-down the E-box3 containing INSM1 promoter region in the Ad-ngn3 transduced PANC-1 cells. This region was not precipitated in the control Ad-LacZ transduced PANC-1 cells (Figure 4B and 4C). Hence, we have conclusively demonstrated that the mechanism for the regulation of the INSM1 gene by ngn3 is via binding the E-box3 element present in the human INSM1 promoter region through the recruitment of the co-activator CBP with a concomitant increase in acetylation of histone H3 and H4.
Figure 4.
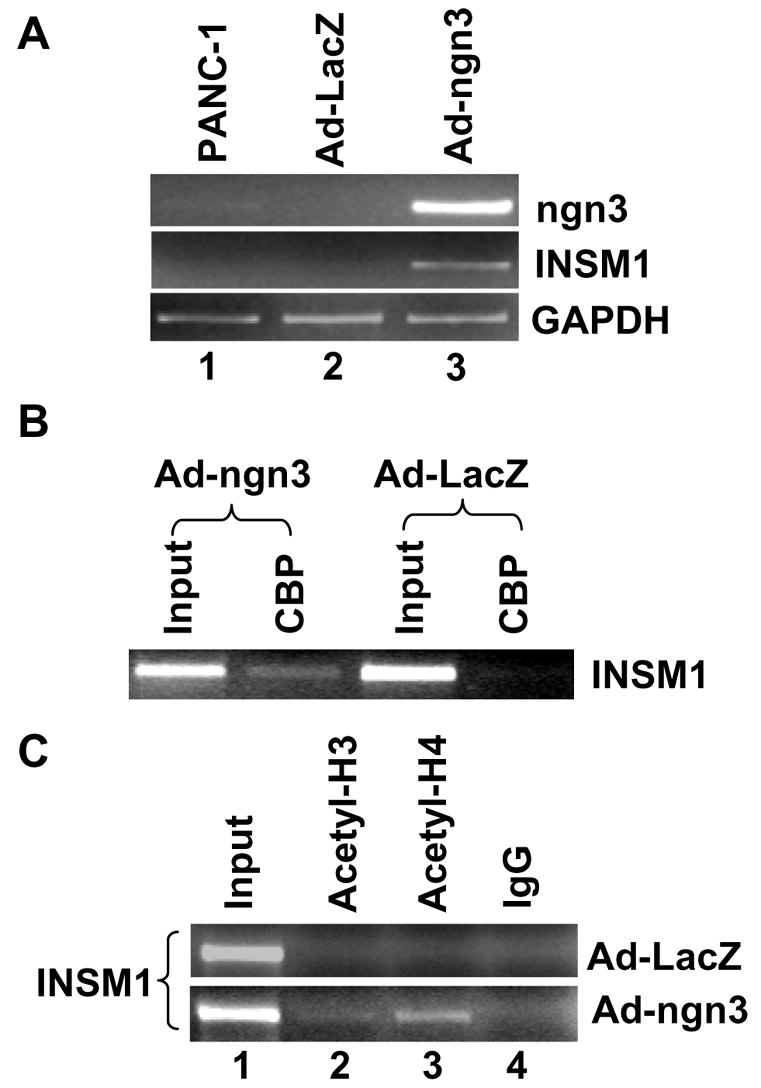
Induction of INSM1 expression in PANC-1 cells and ChIP analyses of CBP and acetyl-histone H3 and H4. (A) Relative RT-PCR analysis of gene expression in Ad-ngn3 transduced PANC-1 cells revealed induction of INSM1 expression as compared to the non-transduced or Ad-LacZ transduced cells. As a control, expression of Ad-ngn3 was performed. GAPDH was included as an internal control for the RT-PCR reaction. (B) ChIP analysis was performed with the Ad-LacZ or Ad-ngn3 transduced PANC-1 cells with an anti-CBP antibody. The CBP antibody could immunoprecipitate the same INSM1 promoter region bound by the ngn3 protein only in ngn3 transduced cells. (C) ChIP analysis was performed with the Ad-LacZ or Ad-ngn3 transduced PANC-1 cells with an anti-acetyl-H3 or anti-acetyl-H4 antibody. Anti-acetyl-H4 and to a lesser degree anti-acetyl H3 antibodies immunoprecipitated the INSM1 promoter only in the Ad-ngn3 transduced PANC-1 cells. Input DNA was included as a positive control (~2.5%) for PCR analysis. Each ChIP experiment was repeated to ensure its reproducibility.
Acknowledgments
This work was supported by funds from the Research Institute for Children, Children's Hospital in New Orleans, and a grant from the NIDDK, National Institutes of Health, DK61436 (to M.S.L.).
Abbreviations
- ngn3
neurogenin 3
- INSM1
insulinoma-associated 1
- CAT
chloramphenicol acetyltransferase
- ChIP assay
chromatin immunoprecipitation assay
- PANC-1
pancreatic ductal carcinoma
- EMSA
electrophoretic mobility shift assay
- bHLH
basic helix-loop-helix
- CBP
CREB-binding protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 2.Wilson ME, Scheel D, German MS. Gene expression cascades in pancreatic development. MechDev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- 3.Habener JF, Kemp DM, Thomas MK. Minireview: transcriptional regulation in pancreatic development. Endocrinology. 2005;146:1025–1034. doi: 10.1210/en.2004-1576. [DOI] [PubMed] [Google Scholar]
- 4.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe DA, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 5.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Neurogenin 3 is required for the development of the four endocrine cell lineages of the pancreas. ProcNatlAcadSciUSA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE, Anderson DJ, Sussel L, Johnson JD, German MS. Expression of neurogenin3 reveals an islet cell precursor popolation in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 7.Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C. The Zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20:2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mellitzer G, Bonne S, Luco RF, Van De Casteele M, Lenne N, Collombat P, Mansouri A, Lee J, Lan MS, Pipeleers D, Nielsen FC, Ferrer J, Gradwohl G, Heimberg H. IA-1 is Ngn3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25:1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto Y, DeSilva MG, Toscani A, Prabhakar BS, Notkins AL, Lan MS. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with zinc-finger DNA-binding motifs. JBiolChem. 1992;267:15252–15257. [PubMed] [Google Scholar]
- 10.Lan MS, Russell EK, Lu J, Johnson BE, Notkins AL. IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res. 1993;53:4169–4171. [PubMed] [Google Scholar]
- 11.Zhu M, Breslin MB, Lan MS. Expression of a novel zinc-finger cDNA, IA-1, is associated with rat AR42J cells differentiation into insulin-positive cells. Pancreas. 2002;24:139–145. doi: 10.1097/00006676-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Breslin MB, Zhu M, Lan MS. NeuroD1/E47 regulates the E-box element of a novel zinc-finger transcription factor, IA-1, in developing nervous system. JBiolChem. 2003;278:38991–38997. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie JP, Cai T, Zhang H, Lan MS, Notkins AL. The zinc-finger transcription factor INSM1 is expressed during embryo development and interacts with the Cbl-associated protein. Genomics. 2002;80:54–61. doi: 10.1006/geno.2002.6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 16.Dumontell E, Laser B, Cpmstant I, Philippe J. Differential regulation of the glucagon and insulin I gene promoters by the basic helix-loop-helix transcription factors E47 and beta 2. JBiolChem. 1998;273:19945–19954. doi: 10.1074/jbc.273.32.19945. [DOI] [PubMed] [Google Scholar]
- 17.Liu WD, Wang HW, Muguira M, Breslin MB, Lan MS. INSM1 functions as a transcriptional repressor of the NeuroD. beta2 gene through the recruitment of cyclin D1 and histone deacetylase. Biochem J. 2006;397:169–177. doi: 10.1042/BJ20051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Notkins AL, Lan MS. Molecular characterization of the promoter region of a neuroendocrine tumor marker, IA-1. Biochem Biophys Res Comm. 1997;236:776–781. doi: 10.1006/bbrc.1997.7054. [DOI] [PubMed] [Google Scholar]
- 19.Huang HP, Liu M, El-hodiri HM, Chu K, Jamrich M, Tsai MJ. Regulation of the pancreatic islet-specific gene beta2 (neruoD) by neurogenin 3. Mol Cell Biol. 2000;20:3292–3307. doi: 10.1128/mcb.20.9.3292-3307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breslin MB, Zhu M, Notkins AL, Lan MS. Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 2002;30:1038–1045. doi: 10.1093/nar/30.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PACF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 22.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. MolCellBiol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 24.Vojtek AB, Taylor J, DeRuiter SL, Yu JY, Figueroa C, Kwok RPS, Turner DL. Akt regulates basic helix-loop-helix transcription factor-coactivator complex formation and activity during neuronal differentiation. MolCellBiol. 2003;23:4417–4427. doi: 10.1128/MCB.23.13.4417-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


