Summary
The Mre11 complex (consisting of MRE11, RAD50, and NBS1/Xrs2) is required for double-strand break (DSB) formation, processing, and checkpoint signaling during meiotic cell division in S. cerevisiae [1–8]. Whereas studies of Mre11 complex mutants in S. pombe and A. thaliana indicate that the complex has other essential meiotic roles [9–11], relatively little is known regarding the functions of the complex downstream of meiotic break formation and processing or its role in meiosis in higher eukaryotes. We analyzed meiotic events in mice harboring hypomorphic Mre11 and Nbs1 mutations which, unlike null mutants, support viability [12–16]. Our studies revealed defects in the temporal progression of meiotic prophase, incomplete and aberrant synapsis of homologous chromosomes, persistence of strand exchange proteins, and alterations in both the frequency and placement of MLH1 foci, a marker of crossovers. A unique sex-dependent effect on MLH1 foci and chiasmata numbers was observed: males exhibited an increase and females a decrease in recombination levels. Thus, our findings implicate the Mre11 complex in meiotic DNA repair and synapsis in mammals and indicate that the complex may contribute to the establishment of normal sex-specific differences in meiosis.
Results
Disruption of Temporal Progression and Synapsis in Meiocytes from Mre11 Complex Mutants
Although histological examination of testis morphology fromMre11 complexhyomorphs indicated that meiogenesis was not grossly disturbed (see Figure S1 in the Supplemental Data available online), subfertility in the mutants [12, 13] was consistent with the hypothesis that Mre11 complex hypomorphism causes perturbations in meiosis. To assess meiotic progression, we determined the distribution of meiotic prophase substages in mutants relative to controls. Oocytes enter meiosis and progress through prophase in a semisynchronous wave during fetal development. Examination of oocytes from Mre11ATLD1/ATLD1 and Nbs1δB/δB females at 17.5– 18.5 days of gestation revealed a significant difference in prophase distribution by comparison with control littermates (Figures S2A and S2B; Mre11ATLD1/ATLD1 χ2 = 122.7, p < 0.0001; Nbs1δB/δB χ2 = 49.4, p < 0.0001). For both mutants, more than 50% of oocytes were at zygotene, whereas fewer than 15% of oocytes from controls remained at zygotene, with the vast majority progressing to pachytene or beyond (for a description of meiotic prophase stages, see Supplemental Experimental Procedures). The paucity of later stages in mutants suggests meiotic delay or arrest at zygotene, the stage in which DSBs are processed and strand exchange intermediates are formed.
Temporal disturbances in meiotic progression were also observed in adult male Mre11 complex hypomorphs. Mre11ATLD1/ATLD1 males exhibited an increase in the proportion of pachytene cells, from40%in controls to 68% in mutants (Figure S2C; χ2 = 109.5, p < 0.0001). Nbs1δB/δB males exhibited a slight but statistically significant increase in the proportion of zygotene cells, from 13% in controls to 20% in mutants (Figure S2D).
Examination of zygotene and pachytene cells from Mre11ATLD1/ATLD1 females (Nbs1δB/δB females were not further examined) and Mre11ATLD1/ATLD1 and Nbs1δB/δB males revealed defects in homologous chromosome synapsis. In Mre11ATLD1/ATLD1 females and males, synaptic defects were evident in 67% (85/126) and 38% (51/134) of pachytene cells, respectively, whereas only 16% (20/123) of control oocytes and 3% (2/68) of control spermatocytes exhibited defects (females: χ2 = 50.9, p < 0.0001; males: χ2 = 28.7, p < 0.0001). The most common aberration in mutant females was partial synapsis of 1 to 3 bivalents (Figure 1A) in cells classified as pachytene; in addition to incomplete synapsis, males commonly exhibited fragmented (Figure 1C) or gapped SCs.
Figure 1. SC Assembly Defects in Prophase Meiocytes from Mre11 Complex Hypomorphic Mice.
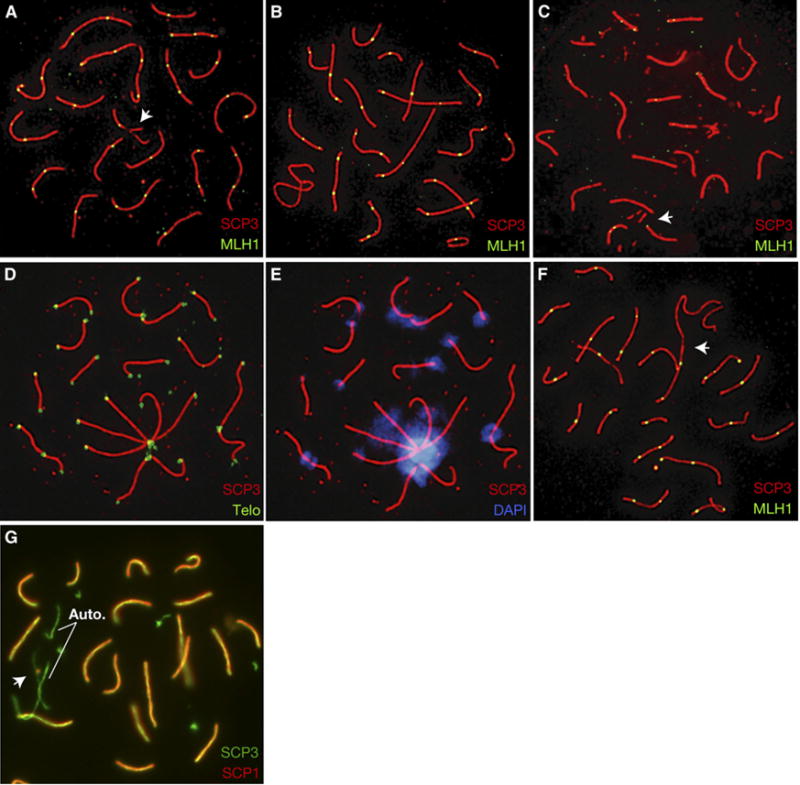
Pachytene cells from Mre11ATLD1/ATLD1 mice exhibit incomplete synapsis (oocyte, [A], arrow), fragmented SCs (spermatocyte, [C], arrow), and SC end associations (oocyte, [D, E]; spermatocyte, [F], arrow). An example of a mutant spermatocyte with normal SC morphology is shown in (B). SC associations result in many overlapping telomere signals ([D], green) and occur at the centromeric ends of SCs ([E], blue). Spermatocytes from Nbs1δB/δB mice exhibit asynapsis in late prophase ([G], auto, indicates autosomal asynapsis; arrow, XY bivalent).
A significant subset of pachytene cells in Mre11ATLD1/ATLD1 mice of both sexes exhibited end-to-end associations between the SCs of nonhomologous chromosomes. In females, associations often involved three or more SCs in a ‘‘pinwheel’’ configuration (19/119 cells versus 3/101 cells in controls, χ2 = 10.2, p < 0.01; Figures 1D and 1E) and occurred exclusively at the centromere-proximal ends of SCs (identifiable by intense centromeric heterochromatin staining; Figure 1E). Because no associations involved distal telomeres, the data do not support the interpretation that these associations result from telomere dysfunction. In males, associations between the X chomosome and autosomal SCs were occasionally observed (Figure 1F) and, like females, involved the centromeric ends of chromosomes.
Nbs1δB/δB males exhibited less severe synaptic aberrations. Nevertheless, 45% (20/44) of cells at the zygotene-pachytene boundary contained an asynaptic bivalent, while all other bivalents were completely synapsed (Figure 1G). In contrast, only 12% (5/39) of similarly staged nuclei from control mice contained a single asynaptic bivalent (χ2 = 10.5, p < 0.01).
Altered Repair of DSBs in Mre11 Complex Mutants
Temporal changes in progression and synaptic aberrations in Mre11 complex hypomorphs suggest significant DSB repair defects. Therefore, we assessed localization patterns of RAD51, an evolutionarily conserved RecA protein required for strand exchange during meiotic DSB repair (see [17, 18–21]). In normal mice, the number of RAD51 foci peaks in leptotene and early zygotene, declining to only a few foci as cells progress to late pachytene and DSBs are resolved [18, 22–24].
We found no difference between Mre11ATLD1/ATLD1 female mice and controls in the number of RAD51 foci at early prophase stages (data not shown), suggesting that the frequency of DSB formation is not significantly altered. However, the mean number of RAD51 foci in pachytene oocytes was significantly increased in mutants (Table S1; U = 1381.0; p < 0.0001). A change in localization pattern was also observed: typically a few RAD51 foci were scattered across all bivalents in controls, whereas mutants exhibited a large number of foci restricted to several SCs (Figures 2A and 2B), giving these bivalents a ‘‘hot’’ appearance. Hot SCs were not necessarily those with synaptic aberrations, as would be expected if these bivalents were temporally out of synchrony with the rest of the cell. Additionally, phosphorylated histone γH2AX (a marker of DSBs; [25]) colocalized to lingering RAD51 foci in mutant oocytes (Figures 2B and 2C), suggesting that these are sites of unrepaired breaks. Specifically, 92 of 93 SCs with 5 or more RAD51 foci (‘‘hot SCs’’) also recruited γH2AX. We observed no evidence of nonhomologous synapsis or exchange, so we do not favor the interpretation that hot SCs represent nonhomologous synapsis.
Figure 2. Persistent RAD51 Localization in Mre11ATLD1/ATLD1 Mice.
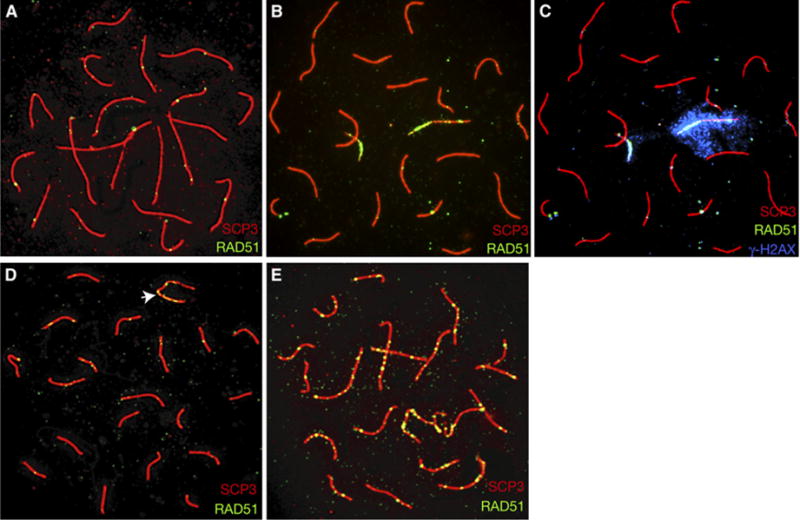
Relative to controls (wild-type oocyte, [A]; wild-type spermatocyte, [D], XY bivalent indicated by arrow), pachytene meiocytes from Mre11ATLD1/ATLD1 mice exhibit prolonged localization of RAD51 foci (mutant oocyte, [B]; mutant spermatocyte, [E]). Persistent RAD51 foci colocalize with γH2AX ([C], same spread as in [B], but overlayed with γH2AX, blue).
Persistent RAD51 foci were also evident in spermatocytes from Mre11ATLD1/ATLD1 males at late pachytene, resulting in an approximate 2.5-fold increase in the mean number of foci (Table S1; U = 315.5; p < 0.05). Additionally, these foci were detected on autosomal SCs, rather than restricted to the XY bivalent as in controls (Figures 2D and 2E). Further, occasional RAD51 foci persisted in some cells at the diplotene stage (data not shown).
Nbs1δB/δB males did not exhibit gross changes in RAD51 foci localization during early prophase. There was, however, a slight, albeit nonsignificant, increase in the proportion of pachytene cells with 1 or 2 remaining RAD51 foci (data not shown), suggesting a similar, though less severe, defect in repair kinetics in this mutant.
Altered Crossover Formation in Mre11 Complex Hypomorphs
The temporal disruption in RAD51 localization raised the possibility of downstream consequences in the recombination pathway, including abnormalities in crossover formation. MLH1 is a MutL family protein required for crossover formation and is commonly used as a marker of exchanges [26–28]. In Mre11ATLD1/ATLD1 females, counts of MLH1 foci in pachytene cells exhibiting complete or nearly complete synapsis revealed a significant decrease in mean foci number (Table 1; t = 5.6, p < 0.0001). In contrast, the average number of MLH1 foci in Mre11ATLD1/ATLD1 and Nbs1δB/δB males was significantly increased (Table 1; t = 4.0, p < 0.0001 and t = 4.3; p < 0.0001, respectively).
Table 1.
Average Number of MLH1 Foci in Pachytene-Stage Meiocytes from Mre11ATLD1/ATLD1 and Nbs1δB/δB Mutant and Control Mice
| Sex | Genotype | Mean # of MLH1 Foci (SD) | Sample Size | Number of Animals |
|---|---|---|---|---|
| Female | Mre11+/+ | 29.2 (4.3) | 62 | 3 |
| Female | Mre11ATLD1/ATLD1 | 24.9 (2.9) | 40 | 5 |
| Male | Mre11+/+ | 24.3 (2.3) | 53 | 2 |
| Male | Mre11ATLD1/ATLD1 | 26.1 (2.9) | 116 | 5 |
| Male | NBS1+/+ | 23.3 (2.3) | 66 | 2 |
| Male | Nbs1δB/δB | 25.1 (2.3) | 55 | 2 |
To further characterize MLH1 patterns in males, the location of foci on single-exchange bivalents was examined. A significant increase in centromere-proximal foci was observed in Mre11 mutants, with a concomitant decrease in the frequency of foci in the most distal segment of the chromosome (Figure 3; χ2 = 20.3, p < 0.005), the preferred region of exchange in wild-type males [29, 30].
Figure 3. A Change in the Position of MLH1 Foci Occurs in Mre11ATLD1/ATLD1 Spermatocytes.
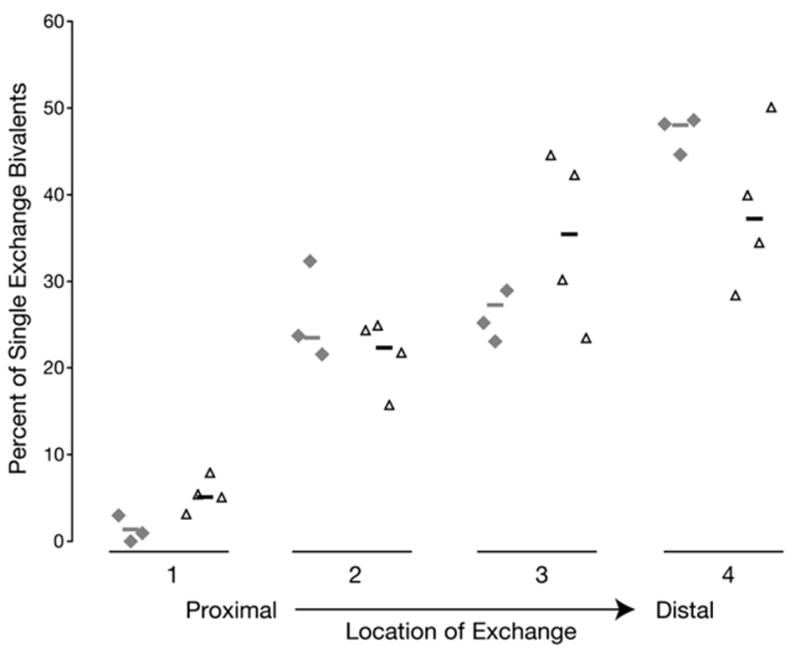
The location of foci relative to the centromere of single exchange bivalents was determined for Mre11ATLD1/ATLD1 mutants in comparison to wild-type mice. The resultant distributions from individual animals are arrayed horizontally along the x axis, with percent of single exchange bivalents scored represented by height of symbol relative to the y axis. Gray diamonds, wild-type males; open black triangles, Mre11ATLD1/ATLD1 males.
To verify the MLH1 data, we analyzed chiasmata in diakinesis meiocytes from Mre11ATLD1/ATLD1 mice. In both sexes, the results paralleled the findings of the MLH1 studies. Mre11ATLD1/ATLD1 females showed a significant decrease in chiasmata (25.1 ± 2.7 versus 27.9 ± 3.2 in controls, t = 3.9, p < 0.01), and males showed a slight, although nonsignificant, increase in chiasmata (23.3 ± 2.2 versus 22.9 ± 1.6 in controls). Failure to achieve statistical significance likely stems from the small number of cells analyzed (technical difficulty prevented obtaining good-quality diakinesis preparations from spermatocytes, also prohibiting analysis of chiasmata location). Additionally, we observed an increase in the frequency of univalents, presumably reflecting defects in synapsis or repair.
Discussion
During the past decade, the phenotypes of knockouts of a variety of meiotic genes (e.g., meiosis-specific cohesins, SC components, and DSB-associated genes) have been reported. A common theme has emerged: synapsis and recombination are codependent processes, and mutations in one pathway disturb the other (reviewed in [31]). Consistent with this, we observed defects in synapsis and persistent RAD51 foci in late prophase in Mre11 complex hypomorphs. However, these mutants also exhibit several unusual meiotic defects, including associations between multiple nonhomolgous SCs and sexually dimorphic effects on crossover formation, indicating novel activities of the complex during the early stages of mammalian meiosis.
Associations between SCs Suggest Defects in the Early Events of Chromosome Pairing
Studies in both yeast and mammals indicate that the earliest events of meiotic recombination set the stage for the subsequent processes of synapsis and crossing over [25, 32, 33]. We found that in the absence of a fully functional Mre11 complex, synapsis was often incomplete or abnormal and frequently involved associations between nonhomologous chromosomes. Early chromosome movements that result in telomere clustering on the nuclear envelope in a ‘‘bouquet’’ are thought to represent a chromosomal reorganization that facilitates homolog synapsis (reviewed by [34, 35]). Intriguingly, all of the end-to-end associations in mutants were restricted to the centromeric ends of chromosomes. Although the role of centromeres in this process remains unknown, this finding suggests that Mre11 complex hypomorphism impedes proper chromosome interactions in early prophase.
Persistence of RAD51 Foci in Pachytene: Evidence of Aberrant Repair
The persistence of strand exchange intermediates observed in both male and female mutants likely reflects alterations in the kinetics of repair. This could be due to a reduction in the rate of DSB processing or an increase in the time required to achieve strand exchange and synapsis. Based on the meiotic DSB-processing activities of the complex in S. cerevisiae, its requirement for meiotic DSB repair in A. thaliana and S. pombe, and its proposed structural activity in bridging DSB ends or sister chromatids [36–38], both are plausible explanations for the persistent strand-exchange phenotype we observed.
Regardless of the underlying mechanism, the most striking feature of the RAD51 studies is that a number of cells were judged to be at mid- to late pachytene on the basis of SC morphology and yet exhibited high numbers of RAD51 foci. Therefore, despite the presence of unrepaired DNA, homologs were able to synapse normally. This does not mean that the Mre11 complex is irrelevant to synapsis, since mutants exhibited significant increases in synaptic defects. Rather, it suggests that, at least in some cells, the synaptic process is uncoupled from DSB repair.
The Mre11 Complex and Crossovers: A Reversal of Sex-Specific Recombination Patterns
The number and position of crossovers are tightly regulated in eukaryotes. Although the mechanism of this control is not well understood, a number of factors appear influential, including DSB distribution, chromatin configuration, and DNA sequence (reviewed by [39– 41]). Our results provide strong evidence that the Mre11 complex participates in this process, with changes in crossover frequency and MLH1 distribution apparent in both Mre11ATLD1/ATLD1 and Nbs1δB/δB animals. Further, the nature of the alterations was unusual in at least two respects. First, in mutant males, exchange events were significantly more frequent than in controls; this is in sharp contrast to previously described meiotic mutations that affect recombination, because virtually all of these involve reductions in recombination [27, 42–44]. Second, the pattern of alterations differed markedly between the sexes, with mutant females exhibiting a decrease in the number of exchanges. As a result, the normal male:female recombination patterns (reviewed in [45]) were reversed, with higher levels in Mre11ATLD1/ATLD1 males than in Mre11ATLD1/ATLD1 females.
Given the essential role of the Mre11 complex in somatic cells, complete elucidation of its meiotic role in mammals must await the production of conditional null alleles. In the interim, however, the meiotic phenotype of Mre11 complex hypomorphs suggests a surprising role in establishment of crossovers. Although defects in DSB repair or changes in the kinetics of repair may indirectly account for the observed changes in crossover formation, we favor a more direct role for the complex in this process. We postulate that the complex contributes to the normal variation in recombination levels between the sexes, possibly via dimorphic associations with chromatin at early prophase stages.
Supplementary Material
Acknowledgments
This work was supported by GM56888, GM59413, and the Joel and Joan Smilow Initiative (J.H.J.P.); HD37502 (P.A.H.); and HD21341 (T.J.H.). We are grateful to H. Hussein, A. Baker, and J. Griswold for superb animal husbandry, J.-C. Kao for technical assistance, and P. Moens, M.A. Handel, T. Ashley, and S. West for kindly providing antisera.
Footnotes
Supplemental Data
Supplemental Data include two figures, one table, and Experimental Procedures and can be found with this article online at http://www.current-biology.com/cgi/content/full/17/4/▪▪▪/DC1/.
References
- 1.Abdullah MF, Borts RH. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2001;98:14524–14529. doi: 10.1073/pnas.201529598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 3.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 4.Nairz K, Klein F. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 7.Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 8.Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 9.Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell. 2004;16:1968–1978. doi: 10.1105/tpc.104.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleuyard JY, Gallego ME, White CI. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma. 2004;113:197–203. doi: 10.1007/s00412-004-0309-1. [DOI] [PubMed] [Google Scholar]
- 12.Williams BR, Mirzoeva OK, Morgan WF, Lin J, Dunnick W, Petrini JH. A murine model of nijmegen breakage syndrome. Curr Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- 13.Theunissen JW, Kaplan MI, Hunt PA, Williams BR, Ferguson DO, Alt FW, Petrini JH. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol Cell. 2003;12:1511–1523. doi: 10.1016/s1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- 14.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 17.Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet Genome Res. 2004;107:201–207. doi: 10.1159/000080598. [DOI] [PubMed] [Google Scholar]
- 18.Ashley T, Plug AW, Xu J, Solari AJ, Reddy G, Golub EI, Ward DC. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma. 1995;104:19–28. doi: 10.1007/BF00352222. [DOI] [PubMed] [Google Scholar]
- 19.Barlow AL, Benson FE, West SC, Hulten MA. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 1997;16:5207–5215. doi: 10.1093/emboj/16.17.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 21.Enomoto R, Kinebuchi T, Sato M, Yagi H, Kurumizaka H, Yokoyama S. Stimulation of DNA strand exchange by the human TBPIP/Hop2-Mnd1 complex. J Biol Chem. 2006;281:5575–5581. doi: 10.1074/jbc.M506506200. [DOI] [PubMed] [Google Scholar]
- 22.Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- 23.Plug AW, Xu J, Reddy G, Golub EI, Ashley T. Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc Natl Acad Sci USA. 1996;93:5920–5924. doi: 10.1073/pnas.93.12.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plug AW, Peters AH, Keegan KS, Hoekstra MF, de Boer P, Ashley T. Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci. 1998;111:413–423. doi: 10.1242/jcs.111.4.413. [DOI] [PubMed] [Google Scholar]
- 25.Mahadevaiah SK, Turner JM, Baudat F, Rogakou E, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 26.Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 27.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 28.Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 30.Lynn A, Schrump S, Cherry J, Hassold T, Hunt P. Sex, not genotype, determines recombination levels in mice. Am J Hum Genet. 2005;77:670–675. doi: 10.1086/491718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawlowski WP, Cande WZ. Coordinating the events of the meiotic prophase. Trends Cell Biol. 2005;15:674–681. doi: 10.1016/j.tcb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 33.Brown PW, Judis L, Chan ER, Schwartz S, Seftel A, Thomas A, Hassold TJ. Meiotic synapsis proceeds from a limited number of subtelomeric sites in the human male. Am J Hum Genet. 2005;77:556–566. doi: 10.1086/468188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bass HW. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol Life Sci. 2003;60:2319–2324. doi: 10.1007/s00018-003-3312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 36.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 37.Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- 38.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 39.Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 40.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 41.van Veen JE, Hawley RS. Meiosis: when even two is a crowd. Curr Biol. 2003;13:R831–R833. doi: 10.1016/j.cub.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 42.Libby BJ, De La Fuente R, O’Brien MJ, Wigglesworth K, Cobb J, Inselman A, Eaker S, Handel MA, Eppig JJ, Schimenti JC. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev Biol. 2002;242:174–187. doi: 10.1006/dbio.2001.0535. [DOI] [PubMed] [Google Scholar]
- 43.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 44.Xu X, Aprelikova O, Moens P, Deng CX, Furth PA. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development. 2003;130:2001–2012. doi: 10.1242/dev.00410. [DOI] [PubMed] [Google Scholar]
- 45.Lynn A, Ashley T, Hassold T. Variation in human meiotic recombination. Annu Rev Genomics Hum Genet. 2004;5:317–349. doi: 10.1146/annurev.genom.4.070802.110217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


