Abstract
The Notch3 N-terminal sequence is conserved across several mammalian species but diverges from the three other Notch proteins. We determined the significance of the N-terminal sequence using deletion mutants. The first 39 amino acids are required for Notch3 receptor expression, processing and functional activity. In contrast, the first 14 amino acids do not appear to enhance function, yet are required to reduce ectopic cytoplasmic expression of Notch3. We screened binding partners for cytoplasmic expressed Notch3 using a yeast two-hybrid assay. Notch3 binds specifically to the proteasome subunit PSMA1, and increased cytoplasmic expression of Notch3 results in inhibition of proteasome activity. Our findings support a multifunctional role for the conserved N-terminal sequence of Notch3: targeting of the protein to the secretory pathway and reduction of cytoplasmic Notch3 expression which may inhibit cytoplasmic functions.
Notch proteins undergo a complex series of proteolytic processing events which are required for signaling. Initially, Notch is targeted to the endoplasmic reticulum (ER) and Golgi, where it undergoes proteolytic processing (at the S1 site) [1]. Upon binding to classic Notch ligands (Delta and Jagged), Notch undergoes extracellular cleavage at the S2 site [2] [3]. The C-terminal product of this event is an intermediate that undergoes further proteolysis within the transmembrane domain (S3 site) to release the Notch intracellular domain (NICD), which translocates to the nucleus and regulates transcriptional activity of target genes, such as the hairy/enhancer of split (HES) genes [4] [5].
The conserved Notch transmembrane receptor family in mammals is composed of four proteins with different regional and temporal expression patterns. Notch signaling is essential during development and for normal homeostasis in many tissues [6]. In the adult, Notch3 is expressed primarily in arterial smooth muscle [7]; pathogenic mutations within epidermal growth factor (EGF) repeats in the extracellular domain of Notch3 receptor result in CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy), a disorder leading to stroke, migraine with aura, and vascular dementia [8].
The purpose of this study was to characterize the N-terminal sequence of Notch3 and to understand its role in normal Notch3 processing and activation. We demonstrate that the N-terminal sequences of Notch3 influence protein processing and function and inhibit ectopic Notch3 expression in the cytoplasm; we show that inhibition of ectopic Notch3 prevents interactions with cytoplasmic proteins, including the proteasome subunit PSMA1.
MATERIALS AND METHODS
Cloning
Deletion constructs Δ14-Notch3 of the 5' end of Notch3 were created by replacing the 5' SalI/ClaI of full length Notch3 with a PCR generated fragment that replaces amino acids 1–14 with a Kozak sequence. The N-terminal deletion construct Δ39-Notch3 was also generated by a PCR approach. Sec-Δ39-Notch3 was generated by creating a new rescriction site in Notch3 and using this site to clone the nearly full insert into pSecTag2 (Invitrogen).
Cell culture and transfection
Cells were transfected using Lipofectamine 2000 (Invitrogen). In all experiments, empty vector DNA was used to bring the total amount of plasmid DNA to final levels (2 ug/12-well or 1 ug/24-well total plasmid DNA).
Western Blots
Notch3 was detected using a 1:1000 dilution of polyclonal rabbit antibody (M-134, Santa Cruz) against the C-terminus of the intracellular domain of Notch3 and IRDye800-labeled anti-rabbit (Rockland, 1:10,000) secondary antibody. The membrane was analyzed and quantitated with an Odyssey imaging system (LI-COR Biosciences).
Ligand-receptor coculture assay
Activation of the Notch3 receptor was induced in a coculture system [9]. In this system, A7r5 cells were transfected with a total of 1 ug of DNA (600 ng Notch3, 200 ng HES-luciferase, and 100 ng Renilla luciferase control plasmid (pRL-TK; Promega), adjusted to 1 ug with empty vector). Eight hours after transfection, cells were cocultured with L cells stably expressing the Notch ligand Jagged1 or Delta1 or with wild type control L cells (a gift from G. Weinmaster). Twenty-four hours after coculture, firefly and renilla luciferase activities were determined using the Dual Luciferase Assay Kit (Promega).
Cytosol extraction
Digitonin was used to selectively permeabilize the plasma membrane of cells which releases cytosolic proteins[10]. Trypsinized cells were permeabilized in 0.005% digitonin for 5 min on ice. Following centrifugation (10,000 rpm for 10 min at 4 °C), the supernatant was harvested and analyzed by western blotting.
Total live cell quantitation
Total live cell quantitation was performed by measuring total lactate dehydrogenase (LDH) using the CytoTox 96 Non-Radioactive Cytotoxicity Assay Kit (Promega). Direct cell counting using trypan blue exclusion was performed in separate experiments to confirm that LDH activity correlated directly with the number of live A7r5 cells in control experiments.
Fas-L induced cell death
At 24 h post transfection, cells were treated with Fas-L (20ng/mL) for 24 h and then stained with PI at 5ug/mL. The number of dead cells (which appeared bright red with characteristic apoptotic nuclei) per high power field (40x) was determined by a blinded investigator.
Yeast two-hybrid screen
The SOS-rescue screen was performed to identify cytoplasmic binding partners that interact with cytoplasmic Notch3[11]. The bait was composed of human SOS fused to Notch3 extracellular domain (amino acids 33-1342); the bait was composed entirely of EGF-repeats. The cdc25A yeast carrying the SOS-Notch3 bait are viable at room temperature, but not at 37°C. When SOS-Notch3 interacts with myristoylated prey, SOS is brought to the membrane, and positive yeast clones can be selected at 37°C. cDNA libraries (Stratagene) from mouse brain and embryo were screened (8.5 and 6 million colonies). Plasmid DNA encoding potential prey cDNA were recovered from yeast colonies and amplified in bacteria for identification and further analysis. Yeast containing an irrelevant cDNA (SOS-MAFB) were used as a negative control to verify binding specificity of the prey.
Coimmunoprecipitation
Cleared cell lysates were mixed with 1 μg of anti-myc (9E10) mouse monoclonal antibody, reacted with 40 μl of packed protein G–agarose (Upstate) and then washed five times with ice-cold PBS (pH 7.4). Captured proteins were analyzed by SDS-PAGE and western blotting.
Proteasome inhibition assay
Ub-FL is a reporter construct which directs the production of a non-hydrolyzable chain of ubiquitin peptides fused to firefly luciferase [12]. In the setting of normal proteasome function, Ub-FL is rapidly degraded by the ubiquitin proteasome system. However, cellular proteasome inhibition stabilizes Ub-FL and increases luciferase activity. 293A cells grown in 96-well plates were transfected with a total of 0.2 μg of DNA per well (170 ng Notch3 variants, 20 ng Ub-FL reporter and 10 ng phRG-TK).
Statistical analysis
All of experiments were repeated three times (or more, when indicated) with reproducible results. Results are presented as the mean ± SD. Statistical significance of differences was assessed using SPSS software (version 10.0). One way ANOVA followed by the Least-Significant-Difference test was used for the comparisons of the means of groups. The level of significance was set at p<0.05.
RESULTS
N-terminal deletions of Notch3 affect processing
The N-terminal sequence of Notch3 contains several novel features not found in other Notch members. Notably, there are strings of arginine, proline, and leucine that are highly conserved between human, mouse, and rat Notch3 (Figure 1A). To test the functions of various regions of the N-terminus of Notch3, we generated three mutant Notch3 cDNAs that were altered at the N-terminus (Figure 1A, bottom). The first clone lacks the N-terminal 14-aa (Δ14-Notch3), which deletes the string of conserved arginine residues and utilizes the second methionine of Notch3 for translational initiation. The second clone lacks the N-terminal 39-aa (Δ39-Notch3), which is predicted to delete the entire predicted signal peptide including the string of leucines close to the predicted site of signal cleavage (P-Sort; R&D Systems, unpublished data). Finally, we replaced the N-terminal 39 amino acids with a consensus secretion signal from IgKappa. (Sec-Δ39-Notch3). The IgKappa sequence is predicted to produce optimal amounts of processed Notch3.
Fig. 1.
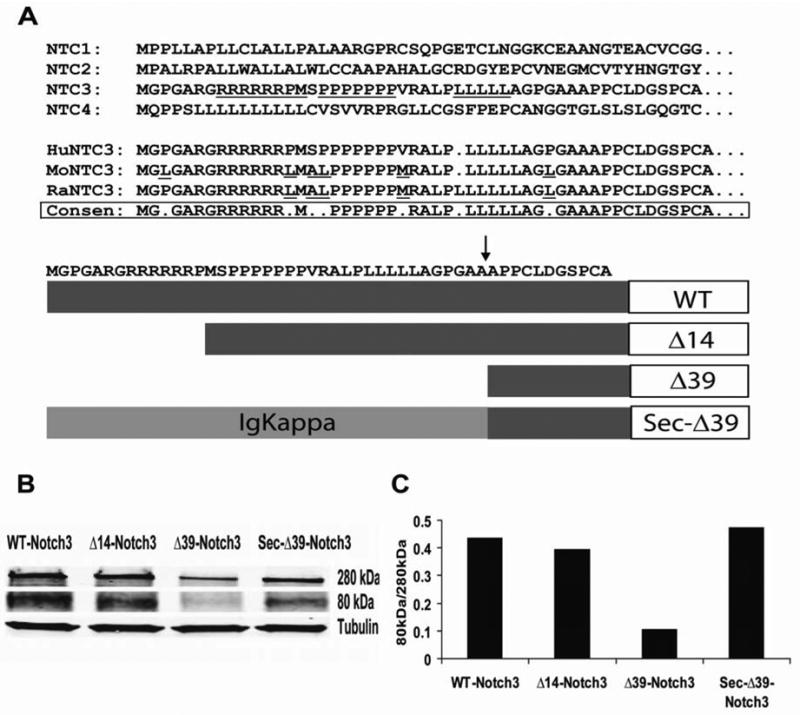
A, Comparison of N-terminal sequences of Notch and evaluation of processing efficiency of deletion mutants. The human Notch3 N-terminus is aligned with Notch3 from mouse and from rat; CONSEN=consensus. The bottom panel shows human Notch3 receptor mutants with N-terminal deletions. An arrow indicates that site predicted to be cleaved by signal peptidase. B, Western-blot analysis of processed vs. unprocessed protein. Upper panel: unprocessed full length Notch3 at 280kDa; middle panel, intracellular domain of Notch3 at 80kDa; lower panel, beta tubulin control for protein loading. C, Processing efficiency of Notch3 proteins. WT, Δ14-Notch3, and Sec-Δ39-Notch3 had similar protein expression and processing, whereas Δ39-Notch3 had notably decreased protein expression and processing.
Western blotting using a C-terminal Notch3 antibody reveals the ratio of intracellular domain (80kDa) and full-length protein (280kDa) [13] [14] and reflects processing efficiency (Figure 1B). The ratios between the 80 kDa and the 280 kDa forms were 0.435, 0.395, 0.105 and 0.474 for WT- Notch3, Δ14-Notch3, Δ39-Notch3 and Sec-Δ39-Notch3, respectively, indicating that the first 14 amino acids have no significant effect on protein expression and processing, but that the first 39 amino acids are critical for protein processing (Figure 1C).
The effect of N-terminal deletions on cytosolic distribution of Notch3
Ectopic localization of Notch3 may have pathological significance, allowing new interactions between cytoplasmic proteins and Notch3. We determined the influence of the N-terminal sequence of Notch3 on cytoplasmic expression of the protein by extracting cells with digitonin, which permeabilizes plasma membrane but leaves intracellular organelles intact. To confirm the purity of digitonin-released cytoplasmic proteins, other subcellular markers, including GM130 (Golgi), HSP60 (mitochondria), VLA alpha-3 (plasma membrane), calnexin (ER), PARP (nucleus) were detected in western blots. Only the cytosol marker tubulin was detected in the digitonin-extracted cytosol (Figure 2A). Cells expressing Δ14-Notch3 contained significantly more cytosolic Notch3 than cells expressing WT-Notch3, which did not express detectable cytoplasmic protein. Cells expressing Δ39-Notch3 had the highest amount of cytosolic Notch3. In the contrast, the insertion of the IgKappa secretion signal sequence in Sec-Δ39-Notch3 inhibited cytosol distribution (Figures 2B and 2C).
Fig. 2.
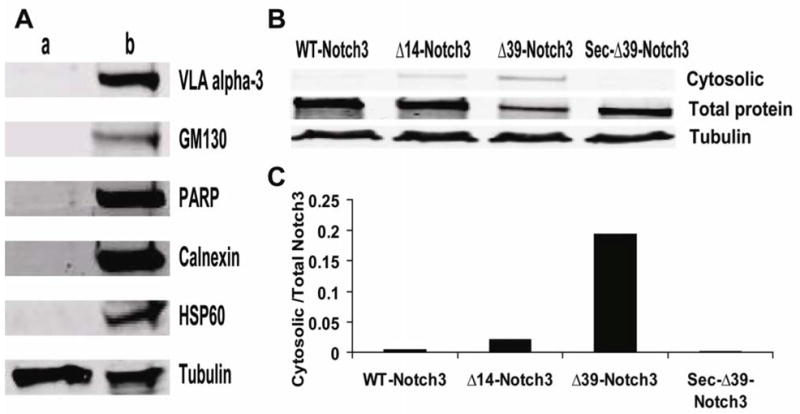
The effect of N-terminal sequences on cytosolic distribution of Notch3. A, Verification of purity of digtonin-extracted cytosol proteins. Left lane (a) cytosolic proteins. Right lane (b): total cell lysates. Proteins were probed for the presence of organelle markers shown (see text for details). B, Notch3 partitioning to the cytosol. Upper panel: digitonin extracted cytosolic Notch3. Middle panel: Notch3 protein from total cell lysates. Lower panel: beta tubulin loading control. C, Ratio of cytosolic to total Notch3. There was an increase in the cytosolic Notch3 from Δ14-Notch3 and Δ39-Notch3 transfected cells, compared with WT-Notch3 and Sec-Δ39-Notch3 transfected cells.
Notch3 N-terminal deletions influence receptor activation
We used a coculture assay to determine the potency of Notch3. As shown in Figure 3A, A7r5 cells exhibit basal stimulation of HES1-luciferase, which is likely mediated by endogenous Notch proteins. Transfection of Notch3 resulted in further increases in reporter activity that was stimulated by Jagged and Delta ligands. Transfected Δ14-Notch3 and Sec-Δ39-Notch3 had ligand stimulated activities that were similar to WT-Notch3; surprisingly, Δ39-Notch3 modestly enhanced HES1-luciferase activity, but the levels were significantly reduced compared to other clones.
Fig. 3.
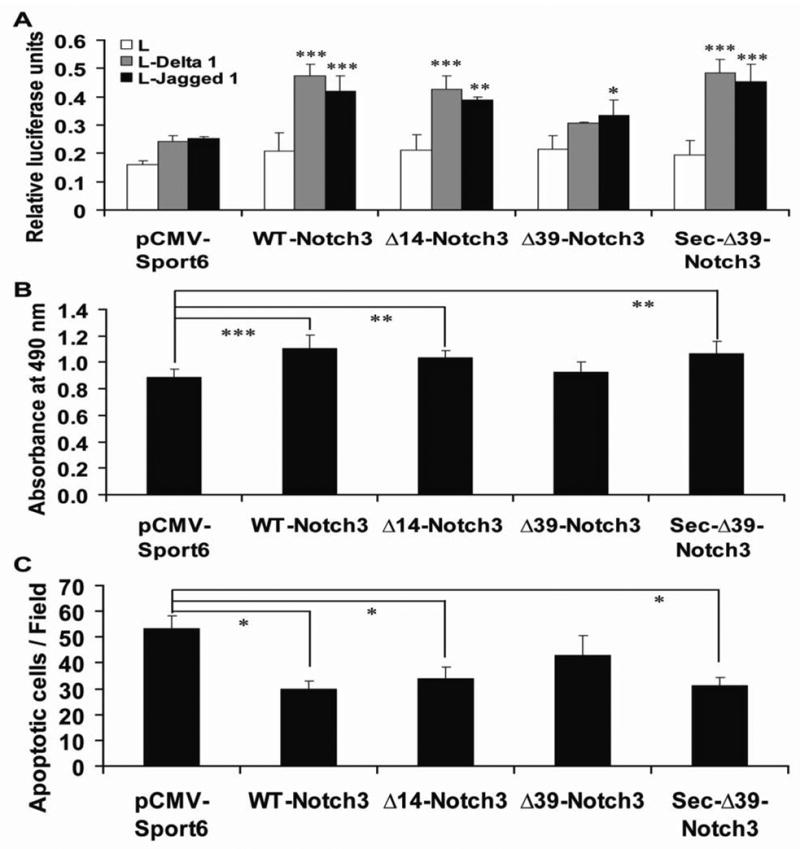
Functional effects of N-terminal deletions of Notch3. A, The activation of the receptor was evaluated using a co-culture assay. A7r5 were cotransfected with Notch3 plasmids, an HES-luciferase reporter, and phRG-TK (transfection normalization control), washed, and then overlayed for 24 hours with L cells or L cells expressing Jagged1 or Delta-like1, which activate HES-luciferase (y-axis) through Notch. B, Influence of the N-terminal deletions on the growth of A7r5 cells. A7r5 cells were transfected and the total cell number after each transfection was determined by quantitation of LDH activity in cell lysates. Cell death assessed by propidium iodide uptake did not change in any transfection. C, The effect of N-terminal deletions on the sensitivity of A7r5 cells to apoptosis. Transfected cells were treated with Fas-L (20ng/mL) for 24 h. Apoptotic cells were stained live using propidium iodide and visualized and quantitated by fluorescent microscopy as described. There was no overall difference in function between WT-Notch3, Δ14-Notch3, and Sec- Δ39-Notch3. However, the function of Δ39-Notch3 was notably impaired in all assays. * p<0.05, ** p<0.01 and ***p<0.001.
Previous studies showed that normal Notch3 signaling promotes the growth of smooth muscle cells[15]. To quantitate this activity of WT and Notch3 mutations, A7r5 cells were transfected with WT-Notch3 or Notch3 mutants. WT-Notch3, Δ14-Notch3 and Sec-Δ39-Notch3 significantly promoted cell growth, while Δ39-Notch3 did not have a significant effect on cell number after transfection (Figure 3B).
Notch3 signaling is a critical determinant of vascular smooth muscle cells survival and vascular structure [16]. Activation of Fas by its ligand (FasL) rapidly induces apoptotic cell death [17]. We thus used the sensitivity to Fas ligand as another measure of functional Notch3 signaling. Whereas WT-Notch3, Δ14-Notch3 and Sec-Δ39-Notch3 effectively protect A7r5 cells against apoptosis induced by Fas-L, Δ39-Notch3 showed a much smaller protective effect (Figure 3C).
Interaction between ectopic cytoplasmic Notch3 and proteasome subunit PSMA1
We were surprised by results comparing WT to Δ14-Notch3. Although there was no difference between the function of WT-Notch3 and Δ14-Notch3, there was a significant increase in the amount of cytosolic Notch3 after digitonin extraction for the Δ14-Notch3 protein. This indicated that the conserved arginine-rich initial 14 amino acids are important for decreasing cytosolic Notch3 expression (Figure 2), but not essential for function (Figure 3). This suggested that there may be important molecular ramifications of ectopic expression of cytoplasmic Notch3.
We performed a genetic screen in yeast to identify proteins that interact with Notch3 in the cytoplasm, using the yeast SOS-rescue screen[11]. The most frequently identified gene encoded PSMA1, a subunit of the 20S proteasome complex. To determine whether PSMA1 and Notch3 could interact in mammalian cells, we performed immunoprecipitations in transfected 293 cells. Notch3 specifically immunoprecipitated with PSMA1, suggesting that the two proteins bind in mammalian cells (Figure 4A).
Fig. 4.
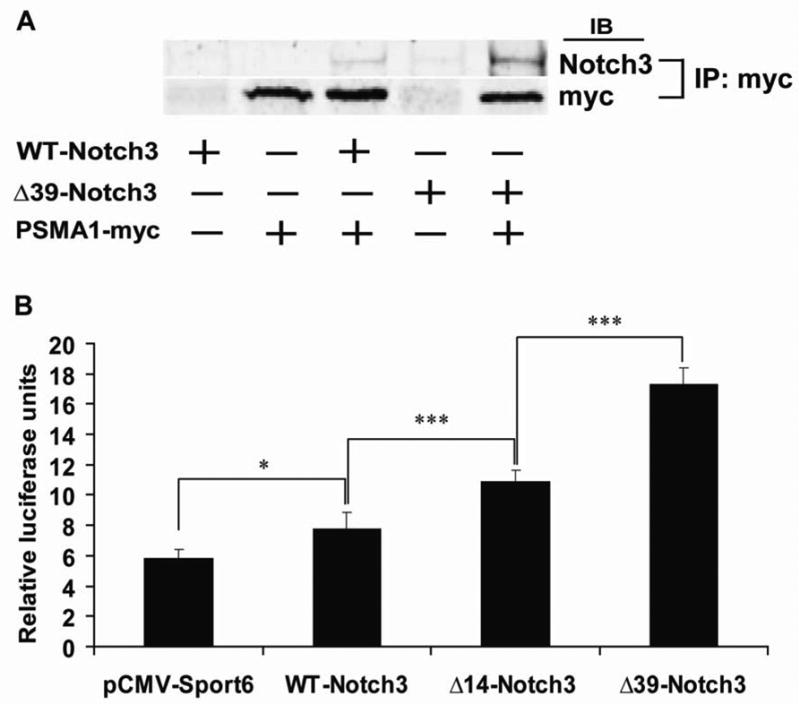
Cytosolic Notch3 interacts with PSMA1 and inhibits proteasome function. A, Coimmunoprecipitation of Notch3 and myc-tagged PSMA1. The plasmids indicated were cotransfected, treated with MG132 for 6 h, and proteins incubated with myc antibodies and pulled down with protein G agarose. Immunoprecipitates were analyzed by immunoblotting with either Notch3 antibodies or myc antibodies. Complexes could only be identified in cell cotransfected with both Notch3 and PSMA1-myc. In addition, only full length Notch3 was identified in immunoprecipitates, indicating that the processed protein could not interact with PSMA1. There was a notable increase in Notch3 immunoprecipitation when progressive amounts of the N-terminus were deleted. B, Cytosolic Notch3 inhibits proteasome function. 293A cells were cotransfected with Notch3 deletions and Ub-FL (with phRG-TK to normalize for transfection efficiency). Luciferase activity (y-axis) is a measure of proteasome inhibition. Cells that were transfected with WT-Notch3 had modestly increased amounts of proteasome inhibition. Progressive deletion of the N-terminus, which showed progressive increases in cytosolic Notch3, results in higher levels of proteasome inhibition. *p<0.05 and ***p<0.001.
To determine whether cytoplasmic Notch3 expression could have functional effects on proteasome function, we used the model substrate Ub-FL which encodes a non-hydroloyzable, polyubiquitin tagged firefly luciferase[12]. When transfected into 293 cells, the reporter reflects proteasome inhibition. There was excellent correlation between the amount of cytoplasmic Notch3 produced (Figure 2), complex formation between PSMA1 and Notch3 (Figure 4A), and the level of proteasome inhibition (Figure 4B).
DISCUSSION
We investigated the role of the atypical, yet conserved, N-terminal sequence of Notch3 in protein processing, localization, and function. The principle findings are: (1) The arginine-rich 14 amino acid leader sequence of Notch3 is required to decrease the amount of ectopic cytosolic Notch3, yet is dispensible for all other tested functions of Notch3. (2) This arginine rich sequence may be important because reduced cytoplasmic expression of Notch3 inhibits physical interactions between Notch3 and the proteasome.
As expected, deletion of Notch3 which excludes the first 39 amino acids of the protein resulted in impaired processing and function loss, confirming that this sequence encodes the complete signal peptide. It is not surprising that exclusion of the first 39 amino acids results in very high cytoplasmic expression of Notch3.
In contrast, the first 14 amino acids of Notch3 are not required for processing, activation, or function of Notch3, yet this sequence inhibits accumulation of cytoplasmic Notch3 production. The conservation of this sequence among mammalian species further supports the importance of inhibition of cytosolic expression by the 14 amino acid sequence. How this sequence inhibits cytoplasmic expression of Notch3 will be a focus of future experiments; possible mechanisms include inhibition of cytoplasmic production of the protein (perhaps by enhancing interactions with the signal recognition particle) or acceleration of degradation of the protein whenever it is ectopically localized to the cytoplasm. The 14 amino acid sequence shares similarity with mitochondrial and nuclear localization signals, and it is conceivable that cytoplasmic protein could be transported to other organelles.
To identify possible deleterious consequences of cytosolic Notch3 expression, we used an interaction trap in yeast and discovered that Notch3 expressed in the cytosol interacts specifically with a proteasome subunit. In addition, proteasome function is progressively inhibited by constructs that result in more cytosolic Notch3 expression. These studies demonstrate that the 14 amino acid stretch of Notch3, which inhibits cytosolic protein localization, may play an important role in prevention of unintended inhibition of proteasome function which could have pathological consequences.
Although WT-Notch3 appears to have very little expression in the cytosol, specific conditions may favor cytosolic expression. One potential mechanism that could result in cytoplasmic expression of EGF-repeat containing ectodomains is retrotranslocation, a process by which misfolded membrane proteins are shuttled across the endoplasmic reticulum for proteasome-mediated degradation. Thus, conditions where there is increased protein misfolding, such as mutations or oxidative stress, could regulate overall proteasome function. Notch3 is a novel example of a protein whose normal location is extracellular, but can potentially be expressed in the cytoplasm. It is indeed likely that many other proteins that are normally extracellular could be misfolded and could interact with and regulate proteasomes, since the EGF repeats of Notch3 (to which PSMA1 binds) are a common structural motif, but this remains to be confirmed experimentally.
The physical interaction between PSMA1 and Notch3 suggests a novel model of proteasome regulation by an ectopic protein. PSMA1 is a subunit that is strategically located at the mouth of the core of the proteasome barrel [18] [19]. While PSMA1 is not part of the catalytic machinery of the proteasome, it likely plays a role in gating entry of proteins into the barrel. Notch3 is a very large protein, and it is conceivable that interactions between PSMA1 and Notch3 may sterically block the core and result in lower overall proteasome function in cells.
Inhibition of proteasome function is thought to play a pivotal role in multiple degenerative conditions, such as Alzheimer’s, Huntington’s, Parkinson’s and Lou Gehrig’s diseases[20] [21], and could potentially play a role in protein accumulation in CADASIL, which is caused by Notch3 mutations that likely lead to misfolding. Increased misfolding is likely to cause an increase in retrotranslocation [22] [23] [24] of the Notch3 protein to the cytoplasm, which subsequently may permit Notch3/proteasome interactions and proteasome inhibition. We are currently evaluating whether CADASIL mutations indeed result in proteasome inhibition. Consistent with this hypothesis, some investigators have detected increased ubiquitinated protein in pathological CADASIL samples[25].
We note that in the yeast two-hybrid screen, we identified both extracellular and intracellular proteins. These proteins bound specifically to the Notch3 ectodomain, raising the possibility that a number of other pathways, in addition to proteasome regulation, could be disrupted by ectopic expression of Notch3. This underscores the potential significance of the Notch3 signal sequence as an added quality insurance mechanism to reduce cytosolic expression of Notch3.
Acknowledgments
We thank Dr. Xiufan Liu (Yangzhou University) for his exceptional support and intellectual guidance. We also thank Drs. Jimo Borjigin, David Piwnica-Worms, and Gerry Weinmaster for reagents and scientific advice and Christina Chae, Susan Lopez, Xiao-Wei Wang, and Geng Geng Yu for technical expertise. This work was supported by NIH (NS41342) and a Burroughs Wellcome Career Award in Biomedical Sciences (1000990).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 3.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 4.Weinmaster G. Notch signal transduction: a real rip and more. Curr Opin Genet Dev. 2000;10:363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 5.Saxena MT, Schroeter EH, Mumm JS, Kopan R. Murine notch homologs (N1-4) undergo presenilin-dependent proteolysis. J Biol Chem. 2001;276:40268–40273. doi: 10.1074/jbc.M107234200. [DOI] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas RMS, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 7.Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, Piga N, Chapon F, Godfrain C, Tournier-Lasserve E. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest. 2000;105:597–605. doi: 10.1172/JCI8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, Maciazek J, Vayssiere C, Cruaud C, Cabanis EA, Ruchoux MM, Weissenbach J, Bach JF, Bousser MG, Tournier-Lasserve E. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 9.Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 10.Afshar N, Black BE, Paschal BM. Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol Cell Biol. 2005;25:8844–8853. doi: 10.1128/MCB.25.20.8844-8853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luker GD, Pica CM, Song J, Luker KE, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- 13.Karlstrom H, Beatus P, Dannaeus K, Chapman G, Lendahl U, Lundkvist J. A CADASIL-mutated Notch 3 receptor exhibits impaired intracellular trafficking and maturation but normal ligand-induced signaling. Proc Natl Acad Sci U S A. 2002;99:17119–17124. doi: 10.1073/pnas.252624099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haritunians T, Chow T, De Lange RP, Nichols JT, Ghavimi D, Dorrani N, St Clair DM, Weinmaster G, Schanen C. Functional analysis of a recurrent missense mutation in Notch3 in CADASIL. J Neurol Neurosurg Psychiatry. 2005;76:1242–1248. doi: 10.1136/jnnp.2004.051854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campos AH, Wang W, Pollman MJ, Gibbons GH. Determinants of Notch-3 receptor expression and signaling in vascular smooth muscle cells: implications in cell-cycle regulation. Circ Res. 2002;91:999–1006. doi: 10.1161/01.res.0000044944.99984.25. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Prince CZ, Mou Y, Pollman MJ. Notch3 signaling in vascular smooth muscle cells induces c-FLIP expression via ERK/MAPK activation. Resistance to Fas ligand-induced apoptosis. J Biol Chem. 2002;277:21723–21729. doi: 10.1074/jbc.M202224200. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M, Burns K, Hahne M, Kennedy N, Kovacsovics M, Tschopp J. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr Biol. 2000;10:640–648. doi: 10.1016/s0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]
- 18.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 19.Powell SR. The cardiac 26S proteasome: regulating the regulator. Circ Res. 2006;99:342–345. doi: 10.1161/01.RES.0000239412.40685.61. [DOI] [PubMed] [Google Scholar]
- 20.Ardley HC, Hung CC, Robinson PA. The aggravating role of the ubiquitin-proteasome system in neurodegeneration. FEBS Lett. 2005;579:571–576. doi: 10.1016/j.febslet.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 21.Layfield R, Lowe J, Bedford L. The ubiquitin-proteasome system and neurodegenerative disorders. Essays Biochem. 2005;41:157–171. doi: 10.1042/EB0410157. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch C, Jarosch E, Sommer T, Wolf DH. Endoplasmic reticulum-associated protein degradation--one model fits all? Biochim Biophys Acta. 2004;1695:215–223. doi: 10.1016/j.bbamcr.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Jarosch E, Lenk U, Sommer T. Endoplasmic reticulum-associated protein degradation. Int Rev Cytol. 2003;223:39–81. doi: 10.1016/s0074-7696(05)23002-4. [DOI] [PubMed] [Google Scholar]
- 24.Plemper RK, Wolf DH. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- 25.Rubio A, Rifkin D, Powers JM, Patel U, Stewart J, Faust P, Goldman JE, Mohr JP, Numaguchi Y, Jensen K. Phenotypic variability of CADASIL and novel morphologic findings. Acta Neuropathol (Berl) 1997;94:247–254. doi: 10.1007/s004010050700. [DOI] [PubMed] [Google Scholar]


