Abstract
HLA-G protein is the functional homolog of Qa-2, the product of the mouse preimplantation embryo development (Ped) gene. Embryos expressing Qa-2 on the cell surface exhibit a faster rate of preimplantation cleavage and preferential survival in utero compared with Qa-2 negative embryos. Qa-2 is glycosylphosphatidylinositol (GPI) linked in the cell membrane. As a result, Qa-2 proteins cluster in cholesterol- and sphingolipid-rich lipid raft microdomains in the cell membrane and can signal via raft-associated intracellular signaling molecules. Using T-cells as a model system, cross-linking of Qa-2 on the cell membrane has been shown to induce proliferation of resting cells. HLA-G, like Qa-2, lacks a cytoplasmic domain capable of transducing signals from the cell surface to the nucleus, but unlike Qa-2, HLA-G has a short 6-amino acid cytoplasmic tail rather than a GPI anchor. In order to test whether HLA-G, like Qa-2, is located in lipid rafts and can act as a signaling molecule we used an HLA-G transgenic mouse system. T-cells were isolated and tested for HLA-G expression by immunofluorescence and for localization of HLA-G in lipid rafts by immunofluorescence and western blotting. Next, the T-cells were cross –linked with anti-HLA-G antibody to test for induction of proliferation. Our novel results show that HLA-G, like GPI-linked Qa-2, is found in lipid rafts in the cell membrane and can act as a signaling molecule to induce proliferation of resting T-cells.
INTRODUCTION
Human embryos fertilized at the same time exhibit a range of developmental rates and preferential survival of faster cleaving embryos [1–3]. These observations echo the phenotype of the Ped (preimplantation embryonic development) gene in mice. Qa-2 protein is the Ped gene product. Mouse embryos expressing membrane-bound Qa-2 protein have an increased rate of preimplantation development and of subsequent embryonic survival than Qa-2 negative mice (reviewed in [4–6]). The functional human homolog of Qa-2 is HLA-G [7–9]. HLA-G mRNA and protein are expressed in preimplantation human embryos [10–13], although one study using only eleven embryos reported no HLA-G mRNA expression [14]. While levels of expression of HLA-G protein varied [15], embryos expressing HLA-G mRNA had significantly faster cleavage rates than embryos lacking message expression [10], recapitulating the Qa-2-mediated Ped phenotype. Production of soluble or shed HLA-G by human embryos has been reported to be associated with improved pregnancy rates after assisted reproductive technologies (ART) and is thought to be clinically predictive of embryo potential [16–20], although it is not yet clear whether presence of HLA-G in embryo culture supernatants is of biological significance or is a general indicator of embryo competence [21, 22].
Both HLA-G and Qa-2 are class Ib MHC proteins with highly homologous extracellular structures [9]. Both molecules lack the canonical MHC class I cytoplasmic tail [7, 23–25], yet they differ in the manner of the insertion of membrane-bound isoforms in the cell membrane. HLA-G has a truncated 6-amino acid cytoplasmic tail [7], while Qa-2 has a glycosylphosphatidylinositol (GPI) linkage to the outer leaflet of the cell membrane [24, 25] and lacks both transmembrane and cytoplasmic residues.
GPI-anchored proteins preferentially localize in lipid rafts in the cell membrane and, indeed, Qa-2 has been shown to be located in lipid rafts [26]. Lipid rafts are liquid ordered subdomains of the plasma membrane. They are enriched in largely saturated sphingolipids and cholesterol, they are resistant to solubilization with non-ionic detergents, and they exhibit light buoyant density in sucrose gradients after ultracentrifugation (reviewed in [27]). Ligation or clustering of GPI-anchored proteins on the cell surface can initiate signal transduction cascades [27–31]. Cross-linking of Qa-2 molecules on mouse T-cells and on embryos induces proliferation of the T-cells and enhances the rate of cleavage of the embryos [32], implicating signaling via lipid raft associated proteins.
The dissimilarity in the tails of the Qa-2 and HLA-G molecules initially suggested that they would not have homologous functions [11]. However, subsequent work indicated that, like the GPI tail of Qa-2, the truncated cytoplasmic tail of HLA-G enables this molecule to associate with lipid rafts [8]. Localization of both Qa-2 and HLA-G in lipid rafts suggests that they could both act as signaling molecules. The effect of cross-linking HLA-G on cell proliferation has not been reported previously and is the subject of this paper. This study used T-cells from a transgenic HLA-G mouse model (HLA-Gtg), in parallel experiments with T-cells from a Qa-2+ mouse strain, to further investigate both the association of HLA-G protein with lipid rafts and the capacity of HLA-G to induce cell proliferation.
MATERIALS AND METHODS
Cell Lines
JEG-3, an HLA-G-expressing choriocarcinoma cell line (ATCC, Manassas, VA) was cultured in Minimal Essential Medium (MEM) (Mediatech, Herndon, PA) supplemented with 10% heat inactivated fetal calf serum (FCS) (Gibco/Invitrogen, Carlsbad, CA), 2 mM L-glutamine, 0.01 mg/mL gentamicin, and 2 mM sodium pyruvate at 37°C and 5% CO2. Cells were detached from the culture flask with trypsin/EDTA (Specialty Media, Phillipsburg, NJ) and washed twice in MEM before use.
EL-4, a Qa-2-expressing mouse lymphoma cell line (ATCC), was cultured in Dulbecco Modified Eagle’s medium (DMEM) (Mediatech) supplemented with 10% heat inactivated horse serum (Gibco), 2 mM L-glutamine, 50 μM 2-mercaptoethanol, 0.01 mg/mL gentamicin, and 2 mM sodium pyruvate at 37°C and 5% CO2.
Mice
C57BL/6(Qa-2+) and CBA/Ca (Qa-2−) mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and bred in the Northeastern University animal facility. HLA-G tg mice, back-bred >20 generations on a CBA/Ca background, were obtained from Dr. Anatolij Horuzsko, Medical College of Georgia, GA, and likewise bred at Northeastern University. Mice were housed in a day-night controlled room (lights on 04:00–18:00 h) with food and water available ad libitum. All experiments were conducted under the guidelines of the NIH in an AAALAC approved facility.
T-cell Enrichment
Mouse spleens were gently mashed through a 70 μm nylon mesh Falcon cell strainer (Fisher Scientific, Pittsburgh, PA) using a rubber-tipped syringe plunger and washed with DMEM producing a cell suspension. To enrich for lymphocytes the cell suspension was layered on Histopaque 1083 (Sigma-Aldrich, St.Louis, MO) pre-warmed to room temperature, and the preparation was spun at 700 x g for 30 minutes at room temperature. The interface lymphocyte cell layer was retrieved, washed in DMEM, and enumerated using a hemocytometer. T-cell enrichment columns were prepared following the manufacturer’s instructions (R&D Systems, Minneapolis, MN) and the eluted cells were re-suspended in serum-free growth medium. Viability was tested by trypan blue exclusion and was always found to be >98%.
Immunofluorescence Staining
1–5 x 10 5 T-cells/well were plated in 96-well V-well plates (Corning Incorporated, Corning, NY). Staining steps were carried out in a total volume of 100 μL 1x phosphate buffered saline (PBS) containing 1% BSA and 0.1% sodium azide (PBSAZ). The cells were washed three times in 200 μL PBSAZ between each staining step. All incubations were done on ice unless otherwise stated. The cells were stained for Qa-2 with 5 μg/mL primary anti-Qa-2 (69H1-9-9, eBiosciences, San Diego, CA) or for HLA-G with 8 μg/mL anti-HLA-G (MEM/G-9, Serotec, Raleigh, NC) antibodies and for lipid rafts with biotinylated CTβ (Molecular Probes, Eugene, OR) at 10 μg/mL for 30 minutes. CTβ labels ganglioside GM1 in lipid rafts. Goat anti-CTβ at 1:100 dilution (Calbiochem, San Diego, CA) was added for an additional 30 minutes. After washing, combined secondary reagents, Alexa Fluor 488 goat F(ab’)2 anti-mouse (Molecular Probes) at 10 μg/mL and a 1:1,000 dilution of streptavidin labeled Qdot 605 (Quantum Dot Corp., Hayward, CA), were added and incubated for 30 minutes. Finally, the cells were fixed in 100 μL 4% paraformaldehyde on ice for 10 minutes. The cells were then plated on cytoslides (ThermoShandon, Pittsburgh, PA) using a cytospin 4 centrifuge (ThermoShandon), mounted using Prolong Gold (Invitrogen/Molecular Probes), covered with #1 coverslips (Fisher Scientific) and allowed to cure overnight before sealing with clear nail polish. Isotype control antibodies were used for staining controls. CBA/Ca T-cells (Qa-2 negative and HLA-G negative) were also used as a negative staining control. Epifluorescence microscopy images were collected using the Keck 3-D fusion microscope [6] with an oil-immersion 100x Total Internal Reflection Fluorescence (TIRF) objective, 1.45 NA, WD 0.17. Images were taken using a model 2.2.1 Spot Camera (Diagnostic Instruments, Sterling Heights, MI) and overlays were processed using Metamorph software (Universal Imaging Corporation, Downingtown, PA). Minimal image cropping was done using Paint Shop Pro™ 7 (Jasc Software Incorporated, Eden Prairie, MN).
Flow Cytometric Analysis
T-cells (5 x 10 5 cells/well) were incubated with anti-Qa-2 (5 μg/mL eBiosciences) or anti-HLA-G (8 μg/mL, Serotec) primary antibody, followed by Goat anti-mouse IgG FITC at 1:800 (ICN/Cappel Aurora, OH) in PBSAZ. Primary and secondary antibodies were incubated for 30 minutes on ice in a total volume of 100 μL. Each incubation step was followed by three washes in 200 μL PBSAZ. Cells were assayed immediately after completion of staining using a FACScan flow cytometer and Cellquest software from Becton Dickinson (Palo Alto, CA). The percentage of T-cells after the purification column was assayed using 1 μg/mL anti-CD3ε (2C11, BD Biosciences Pharmingen, San Diego, CA) primary antibody and 1:200 streptavidin Tricolor (Caltag, Burlingame, CA).
Whole Cell Lysates
The relevanT-cells were washed twice in ice cold PBS. The cells were then resuspended at 1 x 10 6 cells/mL in lysis buffer, (300 mM NaCl, 50 mM Tris-Cl pH 7.6, 0.5% Triton-X 100 and protease inhibitor (Halt Protease Kit, Pierce Biotechnology, Rockford, IL), and incubated on ice 30–45 minutes with light intermittent vortexing. The lysate was then transferred to 1.5 mL microcentrifuge tubes and spun at top speed in a benchtop microfuge for 15 minutes at 4°C. The supernatant was then removed in 25 μL aliquots and stored at −80°C until needed. Immediately prior to western blotting (described below), 2x loading buffer (Jule Inc, Milford, CT) was added to the samples.
Raft Fraction Preparation
A re-suspension of 1.0 x 108 cells in 1 mL lysis buffer containing 20 mM Tris 20 (pH 7.6), 140 mM NaCl, 2 mM EDTA, 0.5 mM NaF, 1mM sodium orthovanadate, 25 mM beta-glycerophosphate, 2 mM sodium pyrophosphate, 2mM benzamidine, 0.5 mM DTT, 0.5% Triton-X 100 and protease inhibitor (Pierce) was incubated on ice for 15 minutes. The lysate was then combined with 1 mL 80% sucrose and the 2 ml 40% sucrose/cell lysate solution was added to a 5ml polyallomer ultracentrifuge tube. A layer of 2 mL 35% sucrose solution was added on top, followed by a top layer of 1 mL 5% sucrose. The tubes were placed in a SW55 Ti swinging bucket rotor (Beckman, Palo Alto, CA) and spun for 18 hours at 100,000 x g in an L5–50B ultracentrifuge (Beckman). At the end of the run, sequential 500 μL aliquots (fractions) were removed from the centrifuge tube, then divided into 400 μL and 100 μL (back-up) aliquots in 1.5 mL microcentrifuge tubes and stored at −80°C. To prepare for western blotting of raft fractions, the protein in the 400 μL fractions was precipitated by adding 800 μL of acetone (pre-chilled to −20°C) and incubated at −20°C overnight before spinning at top speed in a benchtop microfuge at 4 °C for 30 minutes. The supernatant was carefully removed by pipette, and the pellets allowed to air dry at 4 °C. The pellets were then washed twice with 1 ml 80% ice-cold acetone, and finally allowed to air dry at 4 °C for one hour. Twenty μL 2x Tris-Glycine-SDS loading buffer (Jule Inc) were then added to the pellets in preparation for western blotting.
Western Blotting
Samples were boiled for 4 minutes with 2x Tris-Glycine-SDS loading buffer (Jule Inc) and immediately transferred to ice before loading onto 4–20% gradient mini-gels (Pierce). The proteins were separated by SDS-polyacrylamide gel electrophoresis (45 minutes at 100v) and transferred to a Hybond-ECL nitrocellulose membrane (GE Healthcare/Amersham Biosciences, Piscataway, NJ). Membranes were blocked overnight at 4 °C with 5% block solution (GE Healthcare/Amersham), followed by three 10 minute washes with PBS containing 0.1% Tween-20 (PBST) . The membranes were probed with 5 μg/mL anti-Qa-2 (69H1-9-9, eBiosciences) or 2 μg/mL anti-HLA-G (MEM-G/1, Serotec) for 1hour at room temperature on a shaking platform. After three 10 minute washes with PBST, bound antibody was revealed using horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibodies (GE Healthcare/Amersham) by enhanced chemiluminescence (West Pico Kit, Pierce). For lipid raft experiments, after transfer of proteins and overnight blocking, the nitrocellulose membrane was cut at the 25 kD marker, and the lower part of the membrane incubated separately with 10 μg/mL biotinylated CTβ for 1hour at room temperature on a shaking platform. After three 10 minute washes with PBST, bound CTβ was revealed using streptavidin-conjugated horseradish peroxidase (HRP) (GE Healthcare/Amersham) or HRP-conjugated anti –biotin (Cell Signaling Technology Inc, Danvers, MA) by enhanced chemiluminescence (West Pico Kit, Pierce). Longer film exposures revealed GM1 in whole cell lysate controls, but shorter exposures illustrated more clearly the presence of enriched GM1 in raft fractions.
Qa-2 and HLA-G cross-linking on mouse T-cells
T-cells (85–90% CD3ε +) were isolated as described above and 5 x 105 cells/well were cultured in triplicate sets in individual wells of a round bottom 96-well culture plate (Fisher Scientific) in a final volume of 100 μL DMEM supplemented with 10% heat inactivated FCS (Gibco), 2 mM L-glutamine, 50 μM 2-mercaptoethanol, 0.01 mg/mL gentamicin, and 2 mM sodium pyruvate at 37°C and 7.5% CO2. Qa-2+ C57BL/6 T-cells were incubated with anti-Qa-2 antibody (69H1-9-9, eBiosciences). Primary antibody concentration was used in serial dilution from 8 to 0.06 μg/mL. To crosslink the primary antibody, rabbit anti-mouse IgG secondary antibody (ICN/Cappel) was used at varying concentrations (25, 50 and 100 μg/mL) and phorbol myristate acetate (PMA) at a final concentration of 5 ng/mL was added to the cultures. After 48 hours in culture, wells were pulsed with 20 μL of 2 mg/mL MTT (Sigma, St.Louis, MO) to measure cell proliferation. At 72 h, 125 μL of the supernatant were removed, 50 μL of PBS and 150 μL of 0.1 N HCl/isopropanol were added per well and the absorbance at 565 nm was measured in a Synergy HT plate reader (Bio Tek, Winooski, VT). All culture samples were plated in triplicate.
A similar set of experiments were performed on HLA-Gtg T-cells. HLA-Gtg T-cells, at 5 x 105 cells per well, were incubated with purified monoclonal antibody to HLA-G (clone 87G [33], obtained from Dr. D. Geraghty, Fret Hutchinson Cancer Research Center, Seattle, WA). Primary antibody was used at 8 to 0.06 μg/mL, secondary rabbit anti-mouse IgG secondary antibody (ICN/Cappel) was used at 25, 50 and 100 μg/mL, together with PMA at a final concentration of 5 ng/mL. Pulsing of the cells with MTT and measurement of absorbance at 565 nm were as described above.
The specificity of the proliferative response to Qa-2 cross-linking on C57BL/6T-cells was confirmed in control experiments that tested all of the combinations of the components of the experimental set-up. The same controls were tested for the HLA-G T-cell cross-linking model, and, in addition, the control experiments were repeated using T-cells from the Qa-2−, HLA-G− CBA/Ca background mouse strain in which the HLA-Gtg strain had been made. The control data showed that the proliferative response in C57BL/6 and in HLA-Gtg T-cells was induced specifically by cross-linking of Qa-2 and HLA-G respectively, and that all three components, primary antibody, secondary antibody, and PMA were required to induce proliferation (data not shown).
RESULTS
Isolation of CD3ε+ T-cells and confirmation of expression of Qa-2 and HLA-G protein on T-cells from C57BL/6 or HLA-Gtg mice
T-cells were isolated from C57BL/6 (Qa-2+), HLA-G tg (HLA-G+) and CBA/Ca (Qa-2−, HLA-G−) mice for use in several studies: flow cytometry, immunofluorescence microscopy, whole cell lysate western blotting, sucrose gradient ultracentrifugation and lipid raft fraction western blotting, and Qa-2 and HLA-G crosslinking experiments. Figure 1 shows representative data from three independent experiments indicating enrichment of CD3ε+ C57BL/6T-cells (Fig 1a) and CD3ε+ HLA-G tg (Fig 1c) T-cells after histopaque centrifugation and T-cell column enrichment. CD3ε+ cells comprised 85–91% of the T-cell column eluate in each experiment. Enrichment of control CBA/Ca CD3ε+ cells in the same experiments resulted in similar CD3ε+ percentages (data not shown). Confirmation of expression of Qa-2 (open histogram) on C57BL/6 T-cells and of expression of HLA-G (open histogram) on HLA-Gtg T-cells by flow cytometry compared with control CBA/Ca T-cells (shaded histogram) is shown in Figures 1b and 1d, respectively.
FIGURE 1. Qa-2 and HLA-G expression in T-cells from C57BL/6 and HLA-Gtg mice shown by immunofluorescence (FACScan) and western blotting.

a) Post-T-cell enrichment of C57BL/6 CD3ε+ T-cells. Flow cytometric analysis of cell population eluted from T-cell enrichment column showing that 91% of the cells are CD3ε+. Data are representative of three independent experiments where T-cell percentages eluted from columns ranged from 85–91%.
b) Flow cytometric analysis of Anti-Qa-2 staining of C57BL/6 CD3ε+ cells eluted from T-cell column showing that all cells express Qa-2 (open histogram), compared with CBA/Ca T-cells (filled histogram).
c) Post-T-cell enrichment of HLA-Gtg CD3ε+ T-cells. Flow cytometric analysis of cell population eluted from T-cell enrichment column showing that 87% of the cells are CD3ε+. Data are representative of three independent experiments where T-cell percentages eluted from columns ranged from 85–90%.
d) Flow cytometric analysis of Anti-HLA-G staining of HLA-Gtg CD3ε+ cells eluted from T-cell column showing that all cells express HLA-G (open histogram), compared with CBA/Ca T-cells (filled histogram).
e) Western blotting of C57BL/6, CBA/Ca and HLA-Gtg T-cell lysates eluted from T-cell enrichment columns and EL-4 mouse lymphoma cell line lysate with anti-Qa-2 antibody. C57BL/6 T-cell and EL-4 cell lysates are positive for Qa-2, while CBA/Ca and HLA-Gtg T-cell lysates do not express Qa-2.
f) Western blotting of C57BL/6, CBA/Ca and HLA-Gtg T-cell lysates eluted from T-cell enrichment columns and JEG-3 choriocarcinoma cell line lysate with anti-HLA-G antibody. HLA-Gtg T-cell lysate and JEG-3 cell lsate are positive for HLA-G while C57BL/6 and CBA/Ca T-cell lysates do not express HLA-G.
Western blotting experiments reported in Figures 1e and 1f confirm detection of Qa-2 (39 kD) on C57BL/6 T-cells and of HLA-G (45 kD) on HLA-Gtg T-cells. As expected CBA/Ca T-cells are negative for both Qa-2 (Fig. 1e, lane 2) and HLA-G (Fig. 1f, lane 2). Serving as additional negative controls, and confirming that the anti-Qa-2 and anti-HLA-G antibodies used in western blots do not cross-react, C57BL/6 T-cells were probed with anti-HLA-G antibody (Fig. 1f, lane 1) and HLA-Gtg cells were probed with anti-Qa-2 antibody (Fig. 1e, lane 3). EL-4 cell lysate was used as a positive control for Qa-2 blotting (Fig. 1e, lane 4) and JEG-3 cell lysate was used as a positive control for HLA-G blotting (Fig. 1f, lane 4).
Co-localization of Qa-2 and of HLA-G with lipid raft marker GM1 in T-cells from C57BL/6 or HLA-Gtg mice shown by immunofluorescence
Figure 2 shows co-localization of Qa-2 and of HLA-G with lipid raft marker ganglioside GM1 in mouse T-cells detected by fluorescence microscopy. In Figure 2a, T-cells are shown, with Qa-2 labeled by anti-Qa-2 primary antibody and detected with Alexa Fluor 488 goat F(ab’)2 anti-mouse antibody (green) in the first panel, and GM1 labeled by biotinylated CTβ and detected with streptavidin Qdot 605 secondary reagent (red) in the second panel. In the third panel, the images have been overlaid and co-localization of Qa-2 with GM1 is indicated in yellow. In Figure 2b, T-cells are shown, with HLA-G labeled by anti-HLA-G primary antibody and detected with Alexa Fluor 488 goat F(ab’)2 anti-mouse antibody (green) in the first panel, and GM1 labeled by biotinylated CTβ and detected with streptavidin Qdot 605 secondary reagent (red) in the second panel. In the final panel, the images have been overlaid and co-localization of HLA-G with GM1 is indicated in yellow. Figure 2c shows cells incubated with secondary reagents AF488 anti-mouse IgG and streptavidin Q-dot 605 in the absence of primary reagents. We saw no cross-reactivity of anti-Qa-2 antibody against HLA-Gtg T-cells, or of anti-HLA-G against C57BL/6 T-cells (data not shown).
FIGURE 2. Co-localization of Qa-2 and HLA-G with lipid rafts on T-cells from C57BL/6 and HLA-Gtg mice shown by immunofluorescence microscopy.

a) Qa-2 on C57BL/6T-cells is shown in green in the first panel. Primary anti-Qa-2 antibody was detected with AF488-labeled goat anti-mouse IgG. In the second panel, GM1 in lipid rafts was detected using primary biotinylated CTβ followed by streptavidin 605 Qdots. In the third panel, Qa-2 and lipid raft co-localization is shown in yellow in the image overlay. Scale bar 10 μm.
b) HLA-G on HLA-Gtg T-cells is shown in green in the first panel. Primary anti-HLA-G antibody was detected with AF488-labeled goat anti-mouse IgG. In the second panel, GM1 in lipid rafts was detected using primary biotinylated CTβ followed by streptavidin 605 Qdots. In the third panel, Qa-2 and lipid raft co-localization is shown in yellow in the image overlay. Scale bar 10 μm.
c) The first panel shows representative negative control staining of HLA-Gtg T-cells with AF488-labeled goat anti-mouse IgG in the absence of primary anti-HLA-G antibody. The second panel shows representative negative control staining of HLA-Gtg T-cells with streptavidin 605 Qdots in the absence of primary biotinylated CTβ. Scale bar 10 μm.
Enrichment of Qa-2 and of HLA-G in lipid raft fractions of cell lines EL-4 and JEG-3 and in T-cells from C57BL/6 and HLA-Gtg mice shown by ultracentrifugation in sucrose gradients
Localization of Qa-2 and of HLA-G in GM1-enriched lipid rafts was further evaluated by ultracentrifugation of cell lysates in a sucrose gradient, followed by western blotting of the resulting gradient fractions with anti-Qa-2 or anti-HLA-G antibodies. Figure 3a and b show the distribution of Qa-2 in EL-3 cell line and HLA-G in JEG-3 cell line gradient fractions respectively. The fractions are numbered from right to left on the blot with one (top, most buoyant) through 10 (bottom, least buoyant) indicated. A whole cell lysate preparation of EL-4 cells (Fig. 3a) or JEG-3 cells (Fig. 3b) was used as the positive control (+ lane). GM1 western blotting with CTβ was used to confirm the enrichment of Qa-2 and HLA-G in raft fractions (Figure 3c). Longer exposures showed GM1 in the (+) control lanes. These data are consistent with constitutive enrichment of Qa-2 and of HLA-G in membrane fractions containing lipid raft marker GM1 in EL-4 mouse lymphoma and JEG-3 human choriocarcinoma cell lines respectively.
FIGURE 3. Western blots showing enrichment of Qa-2 and HLA-G in lipid raft fractions following ultracentrifugation in sucrose gradients.

Blots are aligned to show protein precipitated from sucrose gradient fractions recovered after ultracentrifugation from the most buoyant (fraction 1) on the right to the least buoyant (fraction 10) on the left. Positive control whole cell lysates are shown in the + lane on the far left.
a) Western blotting of EL-4 raft fractions probed with anti-Qa-2 antibody.
b) Western blotting of JEG-3 raft fractions probed with anti-HLA-G antibody.
c) Western blotting of JEG-3 raft fraction blot probed with CTβ to detect GM1 in raft fractions.
d) Western blotting of C57BL/6 T-cell raft fractions probed with anti-Qa-2 antibody.
e) Western blotting of HLA-Gtg T-cell raft fractions probed with anti-HLA-G antibody.
f) Western Bloting of HLA-Gtg T-cell raft fraction probed with CTβ to detect GM1 in raft fractions.
In subsequent experiments, we repeated the sucrose gradient centrifugation and western blotting experiments in three independent experiments using Qa-2-expressing T-cells from C57BL/6 mice and HLA-G-expressing T-cells from HLA-Gtg mice. Consistent with the results seen in the cell lines, Qa-2 in C57BL/6 T-cells (Figure 3d) and HLA-G in HLA-Gtg T-cells (Figure 3e) are enriched in membrane fractions that are also enriched with GM1 (Figure 3 f). Whole cell lysate preparations of C57BL/6 T-cells or HLA-Gtg T-cells were used as positive controls (+ lane).
Cross-linking of Qa-2 or of HLA-G on resting T-cells from C57BL/6 or HLA-Gtg mice in the presence of PMA induces proliferation
Cross-linking Qa-2 with primary and secondary antibody in the presence of PMA induces resting T-cells to proliferate [34]. All three components of the stimulation protocol, primary antibody, secondary antibody and PMA are required. A typical experiment of cross-linking Qa-2 on resting C57BL/6 T-cells to induce proliferation is shown in Figure 4a. This finding suggested that cross-linking HLA-G might also be able to induce cell proliferation.
FIGURE 4. Proliferation of T-cells from C57BL/6 and HLA-Gtg mice following cross-linking of Qa-2 or HLA-G.
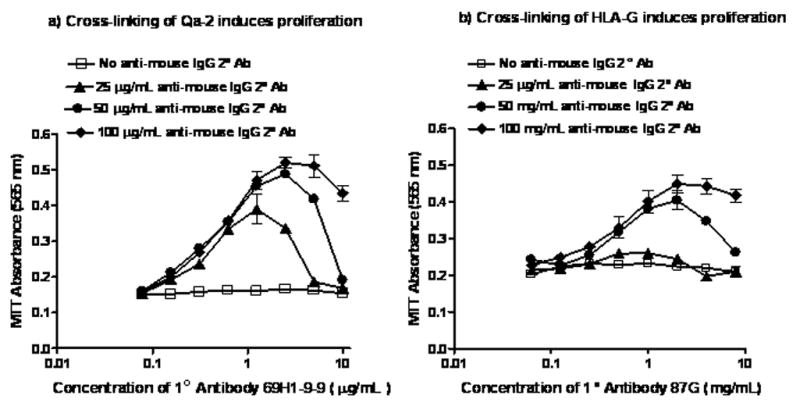
a) Proliferation of C57BL/6 T-cells measured by MTT absorbance at 565 nm (y-axis) in response to serial dilution of anti-Qa-2 antibody from 8 to 0.06 μg/mL (x-axis). Cross-linking secondary anti-mouse IgG antibody concentrations of 25 μg/mL (▴̶), 50 μg/mL (●̶), and 100 μg/mL (♦̶) were used. Results of negative control experiments where no cross-linking secondary anti-mouse IgG antibody was added are also shown (□̶). Samples were cultured and stimulated in triplicate and the data are representative of two independent experiments.
b) Proliferation of HLA-Gtg T-cells measured by MTT absorbance at 565 nm (y-axis) in response to serial dilution of anti-HLA-G antibody from 8 to 0.06 μg/mL (x-axis). Cross-linking secondary anti-mouse IgG antibody concentrations of 25 μg/mL (▴̶), 50 μg/mL (●̶), and 100 μg/mL (♦̶) were used. Results of negative control experiments where no cross-linking secondary anti-mouse IgG antibody was added are also shown (□̶). Samples were cultured and stimulated in triplicate and the data are representative of two independent experiments.
The capacity of HLA-G protein to induce proliferation was tested in similar experiments using purified anti-HLA-G monoclonal antibody clone 87G to cross-link HLA-G on T-cells from HLA-Gtg T-cells. Figure 4b shows the results of experiments titrating primary 87G and cross-linking secondary rabbit anti-mouse IgG, in the presence of 5 ng/mL PMA. As was the case in the anti-Qa-2 experiments, proliferation was seen at a range of primary and secondary antibody concentrations in the anti-HLA-G experiments. In the absence of the secondary antibody, no proliferation was seen at any concentration of primary anti-HLA-G antibody. In the case of the anti-HLA-G cross-linking experiments, however, higher concentrations of secondary rabbit anti-mouse IgG antibody were required to induce proliferation (50 μg/mL in Figure 4b compared with 25 μg/mL with anti-Qa-2 in Figure 4a).
The results of the anti-HLA-G cross-linking experiments showed that cross-linking of HLA-G on the cell surface in the presence of PMA can induce proliferation of T-cells from HLA-tg mice. The response was dependent on the expression of HLA-G, as T-cells from the otherwise genetically identical but HLA-G null CBA/Ca mouse strain did not respond to the same protocol. We did not assay whether the small percentage of non T-cells present in the proliferation experiments (Figure 1) expressed Fc receptor, but the lack of response of T-cells to secondary cross-linking in isotype control assays and in HLA-G null CBA/Ca T-cells appears to rule out involvement of a non-specific Fc-mediated activation mechanism in the proliferation signal observed. In summary, we recapitulated the results of our earlier Qa-2 cross-linking experiments in a similar protocol, and showed that cross-linking of HLA-G resulted in the same functional output-namely induction of proliferation.
DISCUSSION
This study reports the novel finding that membrane-bound HLA-G, the functional human homolog of mouse Qa-2, can localize in lipid rafts and can act as a signaling molecule to induce proliferation upon cross-linking of the protein on the cell surface. Cross-linking of Qa-2 on T-cells [23, 35–38] and on embryos [32] can trigger signaling events that result in cell proliferation, presumably by virtue of the GPI linkage of Qa-2 because it is known that ligation of GPI-anchored proteins on the cell surface can initiate signal transduction cascades. The GPI tail of Qa-2 also allows the molecule to spontaneously incorporate into lipid rafts in cell membranes and ‘painting’ of the protein onto Qa-2 negative embryos has also been shown to increase the rate of preimplantation cleavage of the treated embryos [25]. Functional homology between HLA-G and Qa-2 implies that the truncated six amino acid cytoplasmic tail of HLA-G should also be capable of initiating cellular signaling events.
Lipid rafts containing proteins can dynamically change their size and composition in response to stimuli thus creating cell signaling platforms in the cell membrane and altering protein-protein interactions within the raft microenvironment. Sequestration of proteins into rafts or exclusion of proteins from lipid rafts can facilitate the initiation of signaling cascades, by local cytoplasmic protein tyrosine kinases and phosphatases, via modification of the phosphorylation state of raft-localized proteins. Possession of a canonical intracellular signaling motif by the stimulated protein itself is not a requirement for initiation of a signaling cascade, as accessory proteins in rafts can initiate the cascade in the context of an appropriate stimulus. Examples of proteins lacking canonical cytoplasmic signaling motifs that are nonetheless central to the stimulation of signal transduction in lipid rafts include the T-cell receptor and GPI-linked glial-cell-derived neurotophic factor ( reviewed in [27, 39]).
Some classical Class Ia MHC molecules can induce proliferation on cross-linking (reviewed in [40]) and, interestingly, the signal transduction capacity of human Class Ia MHC molecules is not dependent on the cytoplasmic or transmembrane domains of the proteins. Truncated human Class Ia proteins with as few as four intracytoplasmic amino acids were still capable of activating Jurkat T-cells on cross-linking [41]. We hypothesized that HLA-G, despite its truncated cytoplasmic tail, would therefore also be capable of initiating cellular signaling events upon cross-linking. Alternative use of GPI-anchored versus short cytoplasmic-tailed protein functional homologs in rodents and man is not without precedent in the literature. It has been reported that in carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), a short (10–12 amino acid) cytoplasmic domain isoform in rodent leucocytes has been functionally replaced by a GPI-linked isoform in the human [42]. This study did not investigate whether the short isoforms in rodents were found in lipid rafts, but the location is inferred from a subsequent study showing that CEACAM1 associates with annexin II which is found in lipid rafts [43].
We have previously reported the results of immunofluorescence studies showing that in the JEG-3 human choriocarcinoma cell line HLA-G localizes with lipid rafts containing CTβ-labeled GM1, just as Qa-2 does in the murine lymphoma EL-4 cell line [8]. In the present study, we added to the earlier data by showing that, after ultracentrifugation in sucrose gradients, both HLA-G in JEG-3 cells and Qa-2 in EL-4 cells were enriched in membrane fractions exhibiting lighter buoyant density that were also enriched in GM1 (Figure 3a–c). These experiments confirmed that the truncated cytoplasmic tail of HLA-G enabled it to localize to liquid ordered subdomains of the plasma membrane in the well-established JEG-3 cell line similarly to Qa-2 in the EL-4 line, although they did not provide functional data.
To evaluate functional homology of Qa-2 and HLA-G in terms of their capacity to induce proliferation on cross-linking we decided to use resting T-cells rather than immortalized cell lines which have confounding inherent fast rates of proliferation in culture. Induction of proliferation in C57BL/6 mouse T-cells by cross-linking of Qa-2 had previously been demonstrated [23, 35–38]. Because only 1% of healthy human T-cells express HLA-G protein [44], in order to perform HLA-G cross-linking experiments on T-cells we utilized HLA-G transgenic mice on a Qa-2 negative background [45].
First we confirmed that Qa-2 (Figure 1a, b & e) and HLA-G (Figure 1c, d & f) protein were expressed on T-cells in the C57BL/6 and HLA-Gtg mice by flow cytometry and by western blotting. Next, we showed by immunofluorescence that Qa-2 (Figure 2 a) and HLA-G (Figure 2b) in resting mouse T-cells from C57BL/6 and HLA-Gtg T-cells localize with raft marker GM1 in the cell membrane. Ultracentrifugation of T-cell lysates in sucrose gradients was then used to show that, as in the EL-4 and JEG-3 cell lines, Qa-2 and HLA-G in T-cells was enriched in lighter buoyant membrane fractions that were also enriched in GM1 (Figure 3d–f). Finally, we showed that cross-linking of Qa-2 on C57BL/6 T-cells (Figure 4a) and of HLA-G on HLA-Gtg T-cells (Figure 4b), in the presence of PMA induced proliferation measured by MTT absorbance.
HLA-G is transcribed in many tissues at a level orders of magnitude lower than that of classical class Ia MHC molecules [46], and tissue distribution of HLA-G protein is even more restricted under normal conditions [47]. A role for HLA-G in immunomodulation of several components of the innate and of the adaptive immune response has been proposed. Membrane-bound and soluble isoforms of HLA-G are effective inhibitors of NK-mediated cell lysis [48], by ligation of NK-cell inhibitory ligand KIR2DL4 [49]. This activity is consistent with an important function for HLA-G in protecting the fetus from maternal immune attack at the NK-cell rich maternal/fetal interface and in acceptance of tissue transplants [50].
Soluble HLA-G has also been shown to induce apoptosis in activated CD8+ T and NK cells via a Fas/FasL mediated mechanism [51]. This induction of apoptosis by HLA-G is most likely to apply in the context of local effector mechanisms because soluble classical Class Ia molecules are equally effective inducers of apoptosis and they are present in serum at concentrations three logs higher than that of HLA-G, indicating that the capacity of soluble HLA Class I molecules circulating in serum to induce apoptosis is mainly attributable to soluble Class Ia molecules [51]. The design of our experiments did not address whether the induction of apoptosis was also an effect of HLA-G cross-linking in T-cells, but this is clearly an area of interest for future experiments.
Soluble HLA-G production by allo-specific CD4+ cells from the responder populations in mixed lymphocyte cultures has been reported to suppress the alloproliferative T-cell response [52]. This response may be important in graft acceptance [53], but it is a complicated response as only three of nine allo-combinations tested were capable of producing soluble HLA-G and the same responder cells did not produce soluble HLA-G when in contact with all of the allogeneic stimulator cells tested [52].
The assigned immunomodulatory functions of HLA-G in the context of the innate and adaptive immune response are not easy to reconcile with a role for HLA-G in preimplantation embryo development. In the free-floating preimplantation embryo, which is known to be shielded from cytoxic T-cell lysis by the physical barrier of the zona pellucida [54], it is unlikely that protection from NK or CTL cell lysis by inhibition or apoptosis is the major function of embryonic membrane-bound or soluble HLA-G proteins. Rather, the capacity of HLA-G to induce proliferation in both embryos and T-cells, as described in this paper, is complementary to the immune functions of HLA-G already described.
The murine homolog of HLA-G, Qa-2, is the product of the mouse preimplantation embryo development (Ped) gene, and expression of Qa-2 protein in preimplantation embryos confers a general reproductive advantage. Expression of Qa-2 is not absolutely required for successful reproduction, but a range of advantageous phenotypes including a faster preimplantation cleavage rate and preferential survival of Qa-2+ embryos compared with Qa-2− embryos have been documented (reviewed in [4–6]). Human embryos fertilized at the same time also exhibit a range of developmental rates and preferential survival of faster cleaving embryos [1–3] which suggests that a Ped-like gene exists in humans. HLA-G expression by human embryos has been correlated with enhanced embryo survival [16–20] indicating that HLA-G is the product of the human PED gene. It will be interesting to determine if HLA-G acts as a signaling molecule in embryos from the HLA-Gtg mice in a similar manner in which we have shown in this paper that it can act as a signaling molecule in T-cells.
In conclusion, in this paper we describe a novel function for HLA-G protein in its ability to act as a signaling molecule. This work opens up new vistas for research into the potential mechanisms through which this clinically significant molecule appears to enhance embryo proliferation and survival rates after ART.
Acknowledgments
Supported by National Institutes of Health grants HD 39215, HD 40309 and NSF Grant EEC-9986821. The authors would like to acknowledge and thank Dr.Anatolij Horuzsko, Medical College of Georgia for sharing his HLA-G transgenic mice with us, and Dr. Daniel Geraghty, Fred Hutchinson Cancer Research Center for his kind gift of anti-HLA-G antibody 87G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Van Montfoort AP, Dumoulin JC, Kester AD, Evers JL. Early cleavage is a valuable addition to existing embryo selection parameters: a study using single embryo transfers. Hum Reprod. 2004;19(9):2103. doi: 10.1093/humrep/deh385. [DOI] [PubMed] [Google Scholar]
- 2.Windt ML, Kruger TF, Coetzee K, Lombard CJ. Comparative analysis of pregnancy rates after the transfer of early dividing embryos versus slower dividing embryos. Hum Reprod. 2004;19(5):1155. doi: 10.1093/humrep/deh208. [DOI] [PubMed] [Google Scholar]
- 3.Lundin K, Bergh C, Hardarson T. Early embryo cleavage is a strong indicator of embryo quality in human IVF. Hum Reprod. 2001;16(12):2652. doi: 10.1093/humrep/16.12.2652. [DOI] [PubMed] [Google Scholar]
- 4.Warner CM, Brenner CA. Genetic regulation of preimplantation embryo survival. Curr Top Dev Biol. 2001;52:151. doi: 10.1016/s0070-2153(01)52011-6. [DOI] [PubMed] [Google Scholar]
- 5.Warner CM. Immunological aspects of embryo development. In: Cohen J, Elder K, editors. Human Embryo Evaulation and Selection. Lancaster, U.K: Parthenon Publishing; 2006. [Google Scholar]
- 6.Warner CM, Newmark JA, Comiskey M, De Fazio SR, O’Malley DM, Rajadhyaksha M, Townsend DJ, McKnight S, Roysam B, Dwyer PJ, Dimarzio CA. Genetics and imaging to assess oocyte and preimplantation embryo health. Reprod Fertil Dev. 2004;16(7):729. doi: 10.1071/rd04088. [DOI] [PubMed] [Google Scholar]
- 7.Geraghty DE, Koller BH, Orr HT. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(24):9145. doi: 10.1073/pnas.84.24.9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comiskey M, Goldstein CY, De Fazio SR, Mammolenti M, Newmark JA, Warner CM. Evidence that HLA-G is the functional homolog of mouse Qa-2, the Ped gene product. Hum Immunol. 2003;64(11):999. doi: 10.1016/j.humimm.2003.08.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clements CS, Kjer-Nielsen L, Kostenko L, Hoare HL, Dunstone MA, Moses E, Freed K, Brooks AG, Rossjohn J, McCluskey J. Crystal structure of HLA-G: a nonclassical MHC class I molecule expressed at the fetal-maternal interface. Proc Natl Acad Sci U S A. 2005;102(9):3360. doi: 10.1073/pnas.0409676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jurisicova A, Casper RF, MacLusky NJ, Mills GB, Librach CL. HLA-G expression during preimplantation human embryo development. Proc Natl Acad Sci U S A. 1996;93(1):161. doi: 10.1073/pnas.93.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao W, Brenner CA, Alikani M, Cohen J, Warner CM. Search for a human homologue of the mouse Ped gene. Mol Hum Reprod. 1999;5(6):541. doi: 10.1093/molehr/5.6.541. [DOI] [PubMed] [Google Scholar]
- 12.Jurisicova A, Antenos M, Kapasi K, Meriano J, Casper RF. Variability in the expression of trophectodermal markers beta-human chorionic gonadotrophin, human leukocyte antigen-G and pregnancy specific beta-1 glycoprotein by the human blastocyst. Hum Reprod. 1999;14(7):1852. doi: 10.1093/humrep/14.7.1852. [DOI] [PubMed] [Google Scholar]
- 13.Yao YQ, Barlow DH, Sargent IL. Differential expression of alternatively spliced transcripts of HLA-G in human preimplantation embryos and inner cell masses. J Immunol. 2005;175(12):8379. doi: 10.4049/jimmunol.175.12.8379. [DOI] [PubMed] [Google Scholar]
- 14.Hiby SE, King A, Sharkey A, Loke YW. Molecular studies of trophoblast HLA-G: polymorphism, isoforms, imprinting and expression in preimplantation embryo. Tissue Antigens. 1999;53(1):1. doi: 10.1034/j.1399-0039.1999.530101.x. [DOI] [PubMed] [Google Scholar]
- 15.Jurisicova A, Casper RF, MacLusky NJ, Librach CL. Embryonic human leukocyte antigen-G expression: possible implications for human preimplantation development. Fertil Steril. 1996;65(5):997. [PubMed] [Google Scholar]
- 16.Fuzzi B, Rizzo R, Criscuoli L, Noci I, Melchiorri L, Scarselli B, Bencini E, Menicucci A, Baricordi OR. HLA-G expression in early embryos is a fundamental prerequisite for the obtainment of pregnancy. Eur J Immunol. 2002;32(2):311. doi: 10.1002/1521-4141(200202)32:2<311::AID-IMMU311>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 17.Sher G, Keskintepe L, Nouriani M, Roussev R, Batzofin J. Expression of sHLA-G in supernatants of individually cultured 46-h embryos: a potentially valuable indicator of ‘embryo competency’ and IVF outcome. Reprod Biomed Online. 2004;9(1):74. doi: 10.1016/s1472-6483(10)62113-x. [DOI] [PubMed] [Google Scholar]
- 18.Noci I, Fuzzi B, Rizzo R, Melchiorri L, Criscuoli L, Dabizzi S, Biagiotti R, Pellegrini S, Menicucci A, Baricordi OR. Embryonic soluble HLA-G as a marker of developmental potential in embryos. Hum Reprod. 2005;20(1):138. doi: 10.1093/humrep/deh572. [DOI] [PubMed] [Google Scholar]
- 19.Sher G, Keskintepe L, Fisch JD, Acacio BA, Ahlering P, Batzofin J, Ginsburg M. Soluble human leukocyte antigen G expression in phase I culture media at 46 hours after fertilization predicts pregnancy and implantation from day 3 embryo transfer. Fertil Steril. 2005;83(5):1410. doi: 10.1016/j.fertnstert.2004.11.061. [DOI] [PubMed] [Google Scholar]
- 20.Yie SM, Balakier H, Motamedi G, Librach CL. Secretion of human leukocyte antigen-G by human embryos is associated with a higher in vitro fertilization pregnancy rate. Fertil Steril. 2005;83(1):30. doi: 10.1016/j.fertnstert.2004.06.059. [DOI] [PubMed] [Google Scholar]
- 21.Warner CM, Comiskey M, Clisham PR, Brenner CA. Soluble HLA-G (sHLA-G) a predictor of IVF outcome? J Assist Reprod Genet. 2004;21(9):315. doi: 10.1023/B:JARG.0000045469.08910.1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sargent IL. Does ‘soluble’ HLA-G really exist? Another twist to the tale. Mol Hum Reprod. 2005;11(10):695. doi: 10.1093/molehr/gah196. [DOI] [PubMed] [Google Scholar]
- 23.Robinson PJ, Millrain M, Antoniou J, Simpson E, Mellor AL. A glycophospholipid anchor is required for Qa-2-mediated T-cell activation. Nature. 1989;342(6245):85. doi: 10.1038/342085a0. [DOI] [PubMed] [Google Scholar]
- 24.Tian Z, Xu Y, Warner CM. Removal of Qa-2 antigen alters the Ped gene phenotype of preimplantation mouse embryos. Biol Reprod. 1992;47(2):271. doi: 10.1095/biolreprod47.2.271. [DOI] [PubMed] [Google Scholar]
- 25.McElhinny AS, Exley GE, Warner CM. Painting Qa-2 onto Ped slow preimplantation embryos increases the rate of cleavage. Am J Reprod Immunol. 2000;44(1):52. doi: 10.1111/j.8755-8920.2000.440108.x. [DOI] [PubMed] [Google Scholar]
- 26.Waneck GL, Stein ME, Flavell RA. Conversion of a PI-anchored protein to an integral membrane protein by a single amino acid mutation. Science. 1988;241(4866):697. doi: 10.1126/science.3399901. [DOI] [PubMed] [Google Scholar]
- 27.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 28.Robinson PJ. Signal transduction by GPI-anchored membrane proteins. Cell Biol Int Rep. 1991;15(9):761. doi: 10.1016/0309-1651(91)90031-d. [DOI] [PubMed] [Google Scholar]
- 29.Brown D. The tyrosine kinase connection: how GPI-anchored proteins activate T-cells. Curr Opin Immunol. 1993;5(3):349. doi: 10.1016/0952-7915(93)90052-t. [DOI] [PubMed] [Google Scholar]
- 30.Morgan BP, van den Berg CW, Davies EV, Hallett MB, Horejsi V. Cross-linking of CD59 and of other glycosyl phosphatidylinositol- anchored molecules on neutrophils triggers cell activation via tyrosine kinase. Eur J Immunol. 1993;23(11):2841. doi: 10.1002/eji.1830231118. [DOI] [PubMed] [Google Scholar]
- 31.van den Berg CW, Cinek T, Hallett MB, Horejsi V, Morgan BP. Exogenous CD59 incorporated into U937 cells through its glycosyl phosphatidylinositol anchor becomes associated with signalling molecules in a time dependent manner. Biochem Soc Trans. 1995;23(2):269S. doi: 10.1042/bst023269s. [DOI] [PubMed] [Google Scholar]
- 32.McElhinny AS, Warner CM. Cross-linking of Qa-2 protein, the Ped gene product, increases the cleavage rate of C57BL/6 preimplantation mouse embryos. Mol Hum Reprod. 2000;6(6):517. doi: 10.1093/molehr/6.6.517. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Geraghty DE, Hunt JS. Cytokine regulation of HLA-G expression in human trophoblasT-cell lines. J Reprod Immunol. 1995;29(3):179. doi: 10.1016/0165-0378(95)00942-e. [DOI] [PubMed] [Google Scholar]
- 34.Tabaczewski P, Shirwan H, Lewis K, Stroynowski I. Alternative splicing of class Ib major histocompatibility complex transcripts in vivo leads to the expression of soluble Qa-2 molecules in murine blood. Proc Natl Acad Sci U S A. 1994;91(5):1883. doi: 10.1073/pnas.91.5.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook RG, Leone B, Leone JW, Widacki SM, Zavell PJ. Characterization of T-cell proliferative responses induced by anti-Qa-2 monoclonal antibodies. Cell Immunol. 1992;144(2):367. doi: 10.1016/0008-8749(92)90252-k. [DOI] [PubMed] [Google Scholar]
- 36.Hahn AB, Soloski MJ. Anti-Qa-2-induced T-cell activation. The parameters of activation, the definition of mitogenic and nonmitogenic antibodies, and the differential effects on CD4+ vs CD8+ T-cells. J Immunol. 1989;143(2):407. [PubMed] [Google Scholar]
- 37.Hahn AB, Tian H, Wiegand G, Soloski MJ. Signals delivered via the Qa-2 molecule can synergize with limiting anti-CD3-induced signals to cause T lymphocyte activation. Immunol Invest. 1992;21(3):203. doi: 10.3109/08820139209072259. [DOI] [PubMed] [Google Scholar]
- 38.Houlden BA, Widacki SM, Bluestone JA. Signal transduction through class I MHC by a monoclonal antibody that detects multiple murine and human class I molecules. J Immunol. 1991;146(2):425. [PubMed] [Google Scholar]
- 39.Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275(23):17221. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 40.Skov S. Intracellular signal transduction mediated by ligation of MHC class I molecules. Tissue Antigens. 1998;51(3):215. [PubMed] [Google Scholar]
- 41.Gur H, Geppert TD, Wacholtz MC, Lipsky PE. The cytoplasmic and the transmembrane domains are not sufficient for class I MHC signal transduction. Cell Immunol. 1999;191(2):105. doi: 10.1006/cimm.1998.1417. [DOI] [PubMed] [Google Scholar]
- 42.Singer BB, Scheffrahn I, Heymann R, Sigmundsson K, Kammerer R, Obrink B. Carcinoembryonic antigen-related cell adhesion molecule 1 expression and signaling in human, mouse, and rat leukocytes: evidence for replacement of the short cytoplasmic domain isoform by glycosylphosphatidylinositol-linked proteins in human leukocytes. J Immunol. 2002;168(10):5139. doi: 10.4049/jimmunol.168.10.5139. [DOI] [PubMed] [Google Scholar]
- 43.Kirshner J, Schumann D, Shively JE. CEACAM1, a cell-cell adhesion molecule, directly associates with annexin II in a three-dimensional model of mammary morphogenesis. J Biol Chem. 2003;278(50):50338. doi: 10.1074/jbc.M309115200. [DOI] [PubMed] [Google Scholar]
- 44.Lozano JM, Gonzalez R, Kindelan JM, Rouas-Freiss N, Caballos R, Dausset J, Carosella ED, Pena J. Monocytes and T lymphocytes in HIV-1-positive patients express HLA-G molecule. Aids. 2002;16(3):347. doi: 10.1097/00002030-200202150-00005. [DOI] [PubMed] [Google Scholar]
- 45.Horuzsko A, Antoniou J, Tomlinson P, Portik-Dobos V, Mellor AL. HLA-G functions as a restriction element and a transplantation antigen in mice. Int Immunol. 1997;9(5):645. doi: 10.1093/intimm/9.5.645. [DOI] [PubMed] [Google Scholar]
- 46.Onno M, Guillaudeux T, Amiot L, Renard I, Drenou B, Hirel B, Girr M, Semana G, Le Bouteiller P, Fauchet R. The HLA-G gene is expressed at a low mRNA level in different human cells and tissues. Hum Immunol. 1994;41(1):79. doi: 10.1016/0198-8859(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 47.Carosella ED, Rouas-Freiss N, Paul P, Dausset J. HLA-G: a tolerance molecule from the major histocompatibility complex. Immunol Today. 1999;20(2):60. doi: 10.1016/s0167-5699(98)01387-5. [DOI] [PubMed] [Google Scholar]
- 48.Rouas-Freiss N, Marchal RE, Kirszenbaum M, Dausset J, Carosella ED. The alpha1 domain of HLA-G1 and HLA-G2 inhibits cytotoxicity induced by natural killer cells: is HLA-G the public ligand for natural killer cell inhibitory receptors? Proc Natl Acad Sci U S A. 1997;94(10):5249. doi: 10.1073/pnas.94.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189(7):1093. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carosella ED, Moreau P, Le Maoult J, Le Discorde M, Dausset J, Rouas-Freiss N. HLA-G molecules: from maternal-fetal tolerance to tissue acceptance. Adv Immunol. 2003;81:199. doi: 10.1016/s0065-2776(03)81006-4. [DOI] [PubMed] [Google Scholar]
- 51.Contini P, Ghio M, Poggi A, Filaci G, Indiveri F, Ferrone S, Puppo F. Soluble HLA-A,-B,-C and -G molecules induce apoptosis in T and NK CD8+ cells and inhibit cytotoxic T-cell activity through CD8 ligation. Eur J Immunol. 2003;33(1):125. doi: 10.1002/immu.200390015. [DOI] [PubMed] [Google Scholar]
- 52.Lila N, Rouas-Freiss N, Dausset J, Carpentier A, Carosella ED. Soluble HLA-G protein secreted by allo-specific CD4+ T-cells suppresses the allo-proliferative response: a CD4+ T-cell regulatory mechanism. Proc Natl Acad Sci U S A. 2001;98(21):12150. doi: 10.1073/pnas.201407398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lila N, Amrein C, Guillemain R, Chevalier P, Latremouille C, Fabiani JN, Dausset J, Carosella ED, Carpentier A. Human leukocyte antigen-G expression after heart transplantation is associated with a reduced incidence of rejection. Circulation. 2002;105(16):1949. doi: 10.1161/01.cir.0000015075.89984.46. [DOI] [PubMed] [Google Scholar]
- 54.Ewoldsen MA, Ostlie NS, Warner CM. Killing of mouse blastocyst stage embryos by cytotoxic T lymphocytes directed to major histocompatibility complex antigens. J Immunol. 1987;138(9):2764. [PubMed] [Google Scholar]


