Summary
Heat shock protein 90 (Hsp90) chaperones a key subset of signaling proteins and is necessary for malignant transformation. Hsp90 is subject to an array of post-translational modifications which affect its function, including acetylation. Histone deacetylase (HDAC) inhibitors and knock-down of HDAC6 induce Hsp90 acetylation and inhibit its activity. However, direct determination of the functional consequences of Hsp90 acetylation has awaited mapping of specific sites. We now demonstrate that Hsp90 K294 is acetylated. Mutational analysis of K294 shows that its acetylation status is a strong determinant of client protein and cochaperone binding. In yeast, Hsp90 mutants that cannot be acetylated at K294 have reduced viability and chaperone function compared to wild type or to mutants that mimic constitutive acetylation. These data suggest that acetylation/deacetylation of K294 plays an important role in regulating the Hsp90 chaperone cycle.
Introduction
Heat shock protein 90 (Hsp90) is a molecular chaperone required for stability and function of numerous conditionally activated client proteins, as well as multiple mutated, chimeric, and over-expressed signaling proteins that promote cancer cell growth and survival. Hsp90 is a conformationally flexible protein whose function is modulated by nucleotide (ATP or ADP) dependent association of distinct co-chaperone complexes. Nucleotide exchange and ATP hydrolysis drive the Hsp90 chaperone machine to bind, chaperone, and release client proteins.
Cycling of the Hsp90 chaperone machine is critical to its function. N-terminal Hsp90 inhibitors prevent nucleotide-dependent cycling, stabilize Hsp90’s “open” conformation which is characterized by weaker client protein binding, and ultimately result in the targeting of client proteins to the proteasome (reviewed in (Blagg and Kerr, 2006)). Although ATP binding and hydrolysis have been convincingly implicated in the cycling of the Hsp90 machine, post-translational modification of Hsp90 may also play an important regulatory role. Thus, hyper-phosphorylation of serine and/or threonine residues has been shown to negatively regulate Hsp90 function (Mimnaugh et al., 1995) (Zhao et al., 2001). Recently, the TPR domain-containing phosphatase Ppt1 was shown to associate with and dephosphorylate Hsp90 in yeast (Wandinger et al., 2006). In the absence of Ppt1, Hsp90 was constitutively hyper-phosphorylated and functionally deficient. Although several phosphorylation sites in Hsp90 have been identified, a detailed understanding of their role in Hsp90 function is still emerging (Ogiso et al., 2004).
Reversible acetylation has also been implicated as a regulatory post-translational modification of Hsp90 (Yu et al., 2002). Hsp90 hyper-acetylation has been detected in cells treated with various histone deacetylase inhibitors (HDACi), and the chaperone activity of hyper-acetylated Hsp90 is impaired (Bali et al., 2005b; Nimmanapalli et al., 2003). Although an acetyltransferase responsible for acetylating Hsp90 has yet to be identified, two groups recently identified HDAC6 as an Hsp90 deacetylase (Bali et al., 2005a; Kovacs et al., 2005). However, HDAC6 is unlikely to be the only Hsp90 deacetylase since the initial report identifying Hsp90 hyper-acetylation used the HDACi FK228, which does not inhibit HDAC6 (Blagosklonny et al., 2002; Furumai et al., 2002).
While pharmacologic inhibition and RNA knockdown of HDACs have been very useful in identifying reversible acetylation as a potential regulator of Hsp90 activity, HDACi and/or HDAC knockdown techniques allow study of only the hyper-acetylated chaperone, and the importance of the acetylation state of individual residues of Hsp90 cannot be queried. Furthermore, effects due to hyper-acetylation of proteins other than Hsp90 (including histones) cannot be ruled out. Therefore, identification of specific acetylation site(s) in Hsp90 is crucial to confirm a direct role for this modification in chaperone function. In this study we identify K294 as a discrete acetylation site and we demonstrate the functional consequences in both mammalian cells and in yeast of the acetylation status of this site. Our data identify the acetylation state of K294, a residue conserved in eukaryotic Hsp90, as a key regulator of Hsp90 function.
Results
Hsp90 is Acetylated at More Than One Site
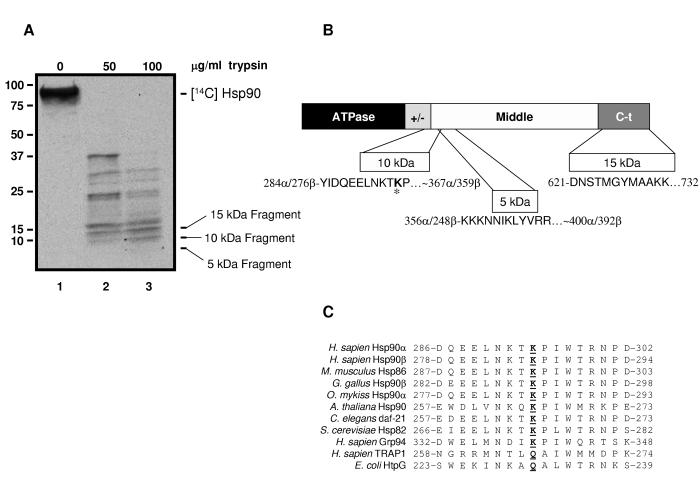
We first sought to identify acetylated domains of Hsp90. We labeled SkBr3 cells with [14C] acetate and immunoprecipitated Hsp90. Following partial proteolysis, acetylated Hsp90 peptides were identified and the smallest peptides were sequenced to locate the domain from which they originated. The 3 smallest peptides detectable by autoradiogram were approximately 15 kDa, 10 kDa, and 5 kDa (Fig. 1A, lane 3). The amino-terminal sequence of each fragment and its location with respect to Hsp90’s domain structure is shown in Figure 1B. The 10 kDa (starting at Y284α/276β) and 5 kDa (starting at K356α/348β) fragments localize to the junction of the charged linker and the middle domain, and to the beginning of the middle domain, respectively. The amino terminal sequence of the 15 kDa fragment begins at D621 and extends to the C-terminus (D732). This is the only fragment that we could clearly identify as α-specific since Ala629 is Met621 in Hsp90β. From these data we conclude that Hsp90 is acetylated at a minimum of two sites.
Figure 1.
Hsp90 is Acetylated in More Than One Domain
(A) SkBr3 cells were labeled with [14C]acetate. Hsp90 was immunoprecipitated from cell lysate, digested with 50 (lane 2) or 100 μg/ml trypsin (lane 3), and fragments were separated by SDS-PAGE and transferred to PVDF membrane. Acetylated bands were detected by autoradiogram. The 3 indicated bands were sequenced.
(B) Diagram of Hsp90 domain architecture showing location and N-terminal sequence of the bands indicated in (A). (ATPase = ATPase domain, +/-= charged linker region, Middle = middle domain, C-t = carboxy-terminal domain; the human Hsp90αsequence was used to determine amino acid numbers). K294 is identified by bolding and an asterisk.
(C) Diagram of K294 relative to Hsp90 domain architecture (domains are as indicated in Fig. 1B). Below is an alignment of the sequence including and around K294 from different organisms and isoforms of Hsp90. K294 or corresponding residues in other organisms/isoforms is underlined and in bold.
Identification of an Hsp90 Acetylation Site at the Junction of the N-terminal and Middle Domains
To unambiguously identify one or more acetylated amino acid(s), we immunopurified Hsp90 from cells treated with trichostatin A (TSA), a broad spectrum HDACi. Hsp90 samples were analyzed for post-translational modification by matrix assisted laser desorption ionization time-of-flight mass spectrometry. The peptide 293α/286β-TKPIWTR-299α/291βwas putatively identified as being acetylated and acetylation of K294α/287β was confirmed by post-source decay (PSD) analysis (data presented in Wang et al. in preparation).
K294α/287βis located at the beginning of the middle domain and is contained within the 10 kDa acetylated peptide fragment identified in Figures 1A and B (indicated in bold with *). The sequence in this region is well conserved from yeast to human Hsp90 (Fig. 1C). However, in TRAP1 (the mitochondrial Hsp90 paralog) and in HtpG (the E. coli Hsp90) K294 is replaced by Q. Interestingly, the region containing K294α/287βhas been reported to be quite antigenic, suggesting that it is exposed to solvent and thus accessible for post-translational modification (Nemoto et al., 1997).
Acetylation of K294 Uniformly Weakens Hsp90 Interaction with Client Proteins
To study the impact of K294 acetylation status on Hsp90αfunction, we made a series of point mutations to mimic acetylated (Q & A) or unacetylated (R) lysine, as has been reported in previous studies (Kim et al., 2006; Wang et al., 2003).
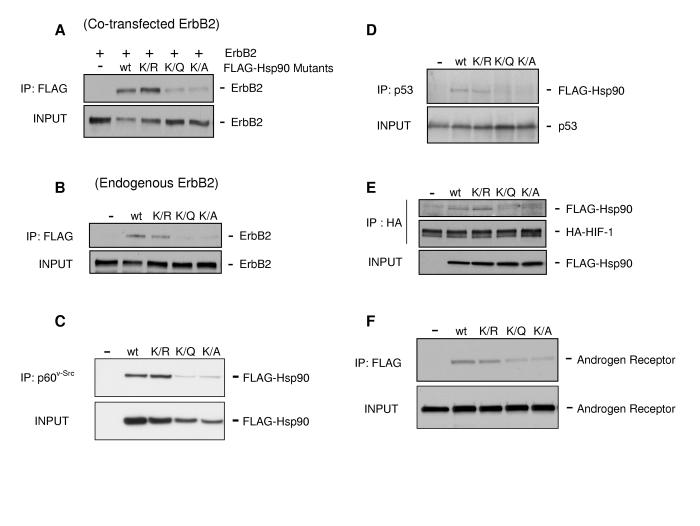
HDACi treatment and HDAC6 knockdown negatively affect Hsp90 interaction with both client proteins and co-chaperones. To ascertain whether the acetylation state of K294 plays a role in client binding, we co-transfected FLAG-tagged WT and mutant Hsp90 constructs together with the Hsp90 client receptor tyrosine kinase ErbB2, and we immunoprecipitated the Hsp90 complex with anti-FLAG antibody. WT and K294R proteins interacted similarly with ErbB2, but both the K294Q and K294A interaction with ErbB2 was dramatically reduced (Fig. 2A). To ensure that this observation was not an artifact of ErbB2 over-expression due to transient transfection, we introduced FLAG-tagged Hsp90 constructs into SkBr3 cells (which express large amounts of endogenous ErbB2) and immunoprecipitated FLAG-Hsp90 complexes as before. Endogenous ErbB2 from SkBr3 cells interacted with WT and K294R but we detected almost no interaction with the K294Q or K294A mutants (Fig. 2B). We obtained nearly identical results with p60v-Src, another Hsp90 client kinase (Fig. 2C).
Figure 2.
Mutation of K294 Affects Hsp90 Interaction with Client Proteins
(A) COS7 cells were co-transfected with ErbB2 (+) and indicated FLAG-Hsp90 constructs. Cells were lysed and FLAG-Hsp90 immunoprecipitates were separated by SDS-PAGE; ErbB2 binding was detected by immunoblotting.
(B) SkBr3 cells were transfected with indicated FLAG-Hsp90 constructs. Hsp90 was immunoprecipitated from cell lysates and ErbB2 binding was determined as in (A).
(C) NIH3T3 cells stably transfected with v-src were transfected with indicated FLAG-Hsp90 constructs. P60v-Src was immunoprecipitated.and associated FLAG-Hsp90 was detected by immunoblotting.
(D) SkBr3 cells were transfected with indicated FLAG-Hsp90 constructs. Endogenous mutant p53 was immunoprecipitated and associated FLAG-Hsp90 was detected as in (C).
(E) COS7 cells were co-transfected with HA-HIF-1αand indicated FLAG-Hsp90 constructs. HA-HIF-1αwas immunoprecipitated and associated FLAG-Hsp90 was detected as in (C).
(F) COS7 cells were co-transfected with androgen receptor and indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and androgen receptor was detected by immunoblotting.
To determine whether this pattern of association was restricted to kinase clients we expanded our analysis to include several transcription factor clients of Hsp90. Mutant and K294A (Fig. 2D & E). Similarly, WT and K294R interacted similarly with the androgen receptor (AR) but K294Q and K294A did not (Fig. 2F). Together these data suggest that acetylation status of K294 significantly and uniformly impacts Hsp90 interaction with a diverse set of client proteins.
Acetylation of K294 Affects Hsp90 Association with Co-chaperones
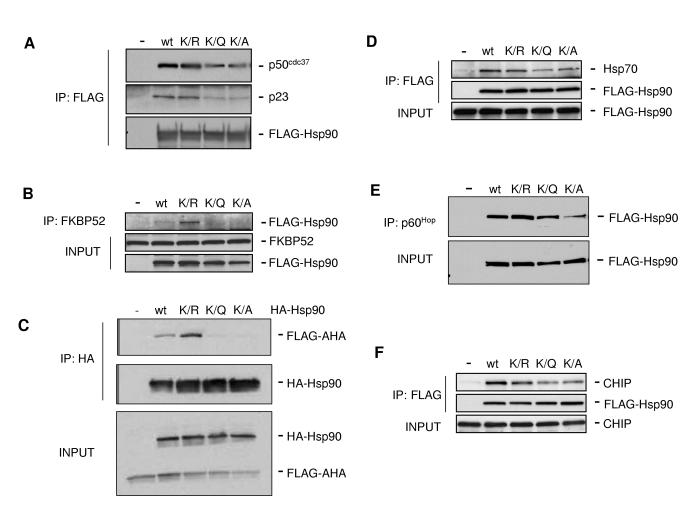
Since Hsp90 functions in dynamic association with other chaperones and co-chaperones we next determined whether acetylation status of K294 affected these interactions. We were able to detect p23 interaction with FLAG-Hsp90 WT and K294R but its interaction with K294Q and K294A was significantly attenuated (Fig. 3A). Similar results were seen for p50cdc37. The 52 kDa FK506 binding protein FKBP52 and the co-chaperone Aha1 also displayed a reduced or undetectable level of interaction with K294Q/A. Strikingly, the K294R mutant displayed a considerably stronger interaction with both FKBP52 and Aha1 than did WT Hsp90 (Fig. 3B, C). Interaction of both K294Q and K294A proteins with Hsp70 and p60Hop was also somewhat diminished relative to WT and K294R (Fig. 3D, E). Finally, we determined the effect of K294 mutation on association of the C-terminus of Hsp70-interacting protein (CHIP) E3 ubiquitin ligase, a co-chaperone implicated in Hsp90 inhibitor-induced degradation of certain clients (Xu et al., 2002). Like p60Hop and Hsp70, CHIP interacted equivalently with WT and K294R but to a lesser degree with K294Q and K294A (Fig. 3F). From these data it is clear that co-chaperones associating with ATP-bound, ADP-bound, and apo-Hsp90 are similarly affected by acetylation state-mimicking K294 mutations.
Figure 3.
Mutation of K294 Affects Hsp90 Interaction with Co-chaperones
(A) COS7 cells were transfected with indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and associated p23 and p50cdc37 were detected by immunoblotting.
(B) COS7 cells were transfected as in (A). FKBP52 was immunoprecipitated and Hsp90 association was detected by immunoblotting.
(C) COS7 cells were co-transfected with FLAG-Aha1 and indicated HA-Hsp90 constructs. HA-Hsp90 was immunoprecipitated and associated FLAG-Aha1 was detected by immunoblotting.
(D) COS7 cells were transfected with indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and Hsp70 binding was detected by immunoblotting.
(E) COS7 cells were transfected with indicated FLAG-Hsp90 constructs. P60Hop was immunoprecipitated and FLAG-Hsp90 binding was detected by immunoblotting.
(F) COS7 cells were co-transfected with CHIP and indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and CHIP binding was detected by immunoblotting.
K294 is Important for Hsp90-dependent Reconstitution of Chk1 Kinase Activity
Since K294Q and A mutants interacted poorly with several client proteins, we predicted that an acetylation-mimicking mutation of K294 would not support client function. To examine this possibility, we utilized a recently established Hsp90-dependent chaperoning system in which purified chaperone components are able to reconstitute the kinase activity of the bacterially expressed Hsp90 client Chk1 (amino acids 1-265) (Arlander et al., 2006). Using this assay, we observed that both WT and K294R were equally capable of activating Chk1 kinase, while the chaperoning ability of K294Q was reduced by 75 percent (Fig. 4A). These data are consistent with a weakened interaction of the K294Q mutant with Chk1 and/or other required co-chaperones such as p50cdc37. To confirm this, we determined the ability of the various purified Hsp90s to interact with both Chk1 kinase and p50cdc37 proteins. As we observed for other kinases in our transfection experiments, K294Q interacted minimally with both Chk1 and p50cdc37 in this purified system (Fig. S1). These data confirm that K294 is important for both client interaction and activity, and that acetylation status of K294 can directly affect the chaperoning of at least one Hsp90-dependent client kinase.
Figure 4.
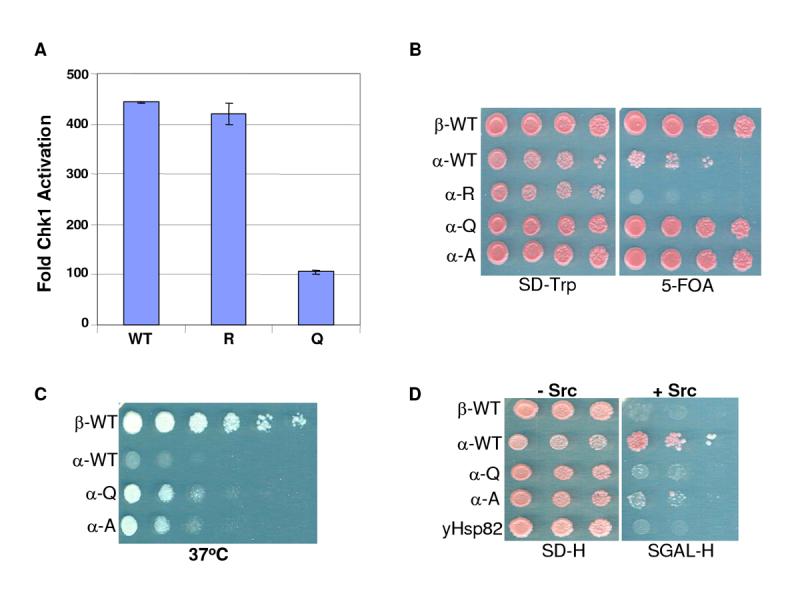
Acetylation Status of Hsp90 K294 Affects Chk1 Kinase Activity In Vitro and Yeast Viability In Vivo
(A) Purified GST-Chk1(1-265) bound to GSH-agarose was incubated with purified WT or indicated mutant Hsp90αproteins together with Hsp70, Ydj, p50cdc37, p60Hop, p23, and CK2. Reactions were washed and precipitates were incubated with [γ-32P]ATP and GST-Cdc25C substrate in kinase buffer. Kinase reaction products were separated by SDS-PAGE and transferred to PVDF membrane. Radiolabeled proteins were imaged and quantitated by phosphorimager. Fold of activation is graphed and standard errors calculated from two experiments.
(B) An S. cerevisiae strain (YKR314) deleted for both genomic copies of yeast HSP90 but carrying both wild type yeast HSP82 (on a URA3 marked plasmid) and human HSP90 constructs (on a low copy CEN-TRP1 marked plasmid) was grown at 30°C to mid-log phase in liquid media. Cultures were adjusted to the same cell density before 5-fold serial dilutions were spotted onto SD-TRP plates either with or without 5-FOA and incubated at 30°C for 3 days before being photographed.
(C) TRP+, ura-colonies were isolated from those strains in (B) which were viable on 5-FOA and grown at 30°C to mid-log phase in liquid YPD media. Cultures were adjusted to the same cell density before 5-fold serial dilutions were spotted onto YPD plates and incubated at 37°C for 3 days before being photographed.
(E) Yeast cells transformed with a GALp-v-src-2μ-HIS3 construct (kindly provided by A. Caplan) and expressing the indicated human HSP90α constructs from CEN-ARS-TRP1 plasmids as the sole source of Hsp90 were grown in liquid SD-HIS media at 30°C to mid-log phase. Cultures were adjusted to the same cell density before serial dilutions were spotted onto either an SD-HIS plate (v-src not expressed; incubated 3 days) or an SGal-HIS plate (v-src is induced; incubated 5 days). Since p60v-Src expression is toxic to yeast, growth indicates that p60v-Src is not functionally expressed while failure to grow on galactose indicates the presence of active p60v-Src protein.
Acetylation-Mimicking Mutants of Human Hsp90αDisplay Enhanced Function in Yeast
We did not observe phenotypic effects of K294 mutation in mammalian cells, presumably because of the high level of endogenously expressed WT Hsp90. Since it would be very difficult to knock down endogenous Hsp90 without also affecting expression of transfected alleles, we addressed the question in S. cerevisiae, a well-characterized in vivo system for studying Hsp90 function (Nathan and Lindquist, 1995; Picard et al., 1990).
We used an S. cerevisiae haploid strain in which the genes encoding both isoforms of endogenous Hsp90 (HSP82 and HSC82) have been disrupted and in which growth is dependent upon HSP82 expressed from a plasmid carrying the URA3 marker, We first transformed this strain (YKR314) with high copy number (2μ) plasmids expressing the various HSP90α alleles and the transformants were re-plated onto media containing 5-Fluoro-orotic acid (5-FOA). We found that neither WT nor K294R human Hsp90αsupported yeast growth under these conditions, while WT Hsp90βand the two Hsp90αmutants mimicking acetylated K294 (K294Q and K294A) supported growth (Fig. S2). Other groups have reported that human Hsp90 functions more effectively when expressed from a low copy number CEN plasmid (Piper et al., 2003) rather than from a high copy number plasmid (Louvion et al., 1998). Thus, we next assessed the ability of all the human Hsp90 constructs to complement yeast when expressed from CEN plasmids. We found that yeast expressing low copy WT Hsp90αwere now viable but grew more slowly than yeast expressing either WT Hsp90βor the hsp90αmutants K294Q or K294A. Yeast expressing human hsp90αK294R as their only source of Hsp90 remained non-viable (Fig. 4B). To further examine the robustness of the various human Hsp90 proteins, we examined the ability of each to support yeast viability under the stress of heat shock (Fig. 4C). WT Hsp90αwas not very effective, suggesting that its function in this yeast strain, even when expressed from a low copy plasmid, is marginal. In contrast, the K294Q and K294A proteins remained functional at elevated temperature, and WT Hsp90βremained the most effective human Hsp90 allele under these conditions. Variations in yeast viability were not due to differential expression of the constructs since they were all expressed at similar levels (not shown).
The Hsp90 client p60v-Src is a promiscuous tyrosine kinase that is toxic to yeast when functionally expressed. This has been used as a phenotypic readout of the ability of various Hsp90 mutants to chaperone p60v-Src in yeast (Nathan and Lindquist, 1995; Xu and Lindquist, 1993). As a final assay for the functionality in yeast of the various human Hsp90 proteins, we examined their ability to chaperone p60v-Src by monitoring cell viability following induction by galactose of a Gal-v-Src construct. Yeast expressing WT hHsp90αremained viable following p60v-Src induction by galactose, indicating an inability to chaperone p60v-src (Fig. 4D). In contrast, hsp90α K294Q and K294A supported p60v-Src function, as did hsp90β as evidenced by the inability of these strains to grow on galactose). Thus, although low-copy expression of WT Hsp9090α can sustain yeast viability, these results together with the heat shock data demonstrate that this protein is not very robust in this yeast strain. However, the K294Q mutant has acquired enhanced functionality in this yeast background, both in its ability to support yeast growth under normal and heat shock conditions, and in its ability to chaperone an exogenously introduced Hsp90 client protein.
Discussion
Hsp90 acetylation dramatically affects its function, but the identity and importance of individual acetylated residues have not been determined. Previous studies have relied on the use of HDACi or HDAC knockdown, but these techniques have their limitations and their direct impact on Hsp90 function is difficult to evaluate. Here we have identified acetylation of a specific lysine residue in the beginning of the middle domain of Hsp90. Both clients and co-chaperones interact within this region, and Hsp90 ATPase activity is regulated by residues in this domain making it an opportune site for post-translational modification (Meyer et al., 2004; Sato et al., 2000). The location of K294 at the junction of the charged linker region and the middle domain places it in a region that is flexible, surface exposed and, based on recent structural data, potentially involved in both intramolecular contacts as well as dynamic domain-domain or protein-protein interactions (Chadli et al., 2000; Huai et al., 2005). In yeast Hsp90, K274 is equivalent to K294 in the human protein. Ali et al. (Ali et al., 2006) point out that in yeast Hsp90 Thr273, Pro275, Try277, Phe292, and Try344 form a hydrophobic pocket for the N-terminal domain Phe200 (213 in human Hsp90α). K274 in yeast Hsp90 is positioned between Thr273 and Pro275, suggesting that its acetylation status might affect protein dynamics.
Conservative mutation of K294 to amino acids that mimic the unacetylated (R) or acetylated (Q or A) state revealed that acetylation status of K294 determines many, but not all, the properties of hyper-acetylated Hsp90. Thus, although hyper-acetylation of Hsp90 following HDACi impairs ATP binding (Murphy et al., 2005; Yu et al., 2002), mutation of K294 did not affect ATP binding (see Fig. S3), suggesting that nucleotide binding is likely regulated by acetylation of other lysine residues within Hsp90. Alternatively, acetylation status of K294 alone may not be sufficient to affect ATP binding. In contrast, consistent with the behavior of hyper-acetylated Hsp90, client and co-chaperone binding was significantly impacted by point mutation of K294. When acetylation of this residue was mimicked, a consistent and substantial decrease in client protein binding was seen. Although a similar trend occurred when co-chaperone interactions were studied, some co-chaperones were less sensitive to K294 substitutions, most notably p60hop, Hsp70, and CHIP. FKBP52 and Aha1 were most dependent on acetylation status of K294, even though they interact with different regions of Hsp90. Additionally, K294 acetylation is a nucleotide-independent determinant of p23 binding to Hsp90. Thus, the acetylation status of K294 may propagate complex allosteric information throughout the chaperone that is independent of, but perhaps synergistic with nucleotide binding. Structural analysis of K294 mutant proteins will be necessary to explore this possibility further.
The functional consequences for Hsp90 chaperone activity of K294 acetylation status were confirmed by the Chk1 in vitro reconstitution assay. Mutation of K294 to mimic the acetylated state significantly abrogated Hsp90’s ability to activate purified Chk1 kinase. This finding is similar to a recent study of Murphy et al. (2005). They found that Hsp90 from cells with HDAC6 knocked down was not able to form a stable complex with glucocorticoid receptor but could only participate in dynamic association with this client protein. Mutation of K294 may allow a similar dynamic (e.g., weak) interaction with clients; however, activation of the Chk1 fragment may require a more stable association with Hsp90.
To examine affects of mutating Hsp90 K294 in an in vivo system, we used S. cerevisiae where both endogenous HSP90 genes were deleted and viability was supported by an episomally carried version (HSP82) that was shuffled with plasmids expressing various alleles of human HSP90. When we expressed WT Hsp90αfrom a high copy plasmid, yeast were unable to grow. However, growth was rescued by Hsp90α K294Q, and Hsp90αK294A (but not by Hsp90αK294R). In a study of Hsp90α expression in yeast, MacLean et al. (MacLean et al., 2005) reported that Hsp90α expressed from a low copy CEN plasmid supported growth, although not as effectively as either Hsc82 or Hsp90β. Our data confirm that there is a difference between Hsp90αand βfunction in yeast and that this difference becomes more pronounced when these proteins are expressed from high copy plasmids. Importantly, this difference is mitigated by acetylation-mimicking mutation of K294 in Hsp90α. When we expressed human HSP90αalleles using CEN plasmids we found, as reported by MacClean et al. (MacLean et al., 2005), that WT Hsp90αweakly supported yeast growth. However, yeast carrying hsp90αK294R on CEN plasmids remained non-viable. In contrast, the robustness of both acetylation-mimicking Hsp90αmutants was further enhanced by low copy expression. High copy plasmids are maintained at approximately 20-50 copies per cell, while CEN plasmids are typically present at 1-2 copies per cell. Since expression levels of mutant and WT Hsp90αproteins from both plasmids were internally comparable (not shown), our data show that K294 acetylation enhances the ability of highly expressed Hsp90αto support yeast growth, perhaps by weakening association with yeast client proteins. In this yeast strain K294 acetylation may be inefficient or its de-acetylation may be too active, so that WT Hsp90αmay bind too tightly to its yeast clients to be fully functional. Although reduced expression allows WT Hsp90αto complement yeast, K294 acetylation (K294Q) remains decidedly beneficial. The fact that Hsp90αK294R is unable to complement yeast even when expressed at low copy suggests that the acetylation state of this residue is critical for its chaperone activity in this system. Inability to acetylate K294 (K294R) may further strengthen Hsp90/client protein interaction to the point where the Hsp90 chaperone machine can no longer cycle.
The p60v-Src and heat shock data support the hypothesis that the robustness of Hsp90αin yeast is dramatically improved by acetylation mimicking mutation of K294 (and worsened by de-acetylation mimicking mutation). Our results also emphasize the difference in chaperone activity in this yeast strain between Hsp90α and β,suggesting that this system may be useful for investigating the unique functional capabilities of each isoform, as well as the underlying molecular determinants regulating their activity. While Hsp90 acetylation in yeast has been little studied to date, in preliminary experiments we have detected steady-state acetylation of yeast Hsp82 that is sensitive to HDACi treatment (not shown). Given the vast number of genetically characterized strains available, exploration of how Hsp90 acetylation/de-acetylation is regulated in yeast will no doubt be highly informative.
In summary, our data are consistent with a model in which acetylation of K294 decreases affinity for most clients and certain co-chaperones while deacetylation increases these interactions. Through its impact on Hsp90 complex dynamics, reversible acetylation of K294, and potentially of other sites in the chaperone, likely provides an additional layer of physiologic control of Hsp90 function in response to environmental signals. Untangling the interplay of various post-translational modifications influencing this process is a challenging but necessary step toward understanding the regulation of this critical chaperone.
Experimental Procedures
Cell Lines and Culture Conditions
COS7, SkBr3, MCF7, and NIH3T3 cells were cultured in a temperature controlled incubator (37°C and 5% CO2) in DMEM medium supplemented with 10 mM HEPES pH 7.0, 10% fetal bovine serum, 2 mM Glutamine, 1 mM Sodium Pyruvate, and nonessential amino acids (Biosource/Invitrogen). For androgen receptor transfection, cells were grown in DMEM medium supplemented with charcoal-stripped fetal bovine serum (Hyclone). NIH3T3 cells stably expressing v-src have been described previously (Sartor et al., 1992; Whitesell et al., 1994).
[14C] Acetate Labeling, Immunoprecipitation, Partial Proteolysis, and Edman Degradation Sequencing
SkBr3 cells were labeled with 80 μCi[14C] sodium acetate (Amersham) for 20 hours and lysed in buffer containing 50 mM Tris-HCl pH7.5, 100 mM NaCl, 1 mM EDTA (Biosource/Invtrogen), 1% NP40, 2 mM Na3VO4, 30 mM NaF, 10 mM sodium butyrate (Sigma), and protease inhibitors (Roche). Protein concentration was determined by BCA protein assay (Pierce), and Hsp90 was immunoprecipitated with anti-Hsp90 antibody (Affinity Bioreagents). The amount of Hsp90 purified was estimated by silver staining. Partial proteolysis of Hsp90 was adapted from Hartson et al. (Hartson et al., 1999). After 4 washes with lysis buffer the immuno-resin was equilibrated by washing 3 times in digestion buffer (10 mM Tris-HCl pH7.5, 100 mM NaCl, 4 mM CaCl2, 0.1 mM EDTA) and resuspended in a volume equal to half the estimated bead volume. Approximately 30 The reactions were incubated on ice for 6 minutes and stopped by boiling in SDS-PAGE sample buffer. Samples were separated by SDS-PAGE and transferred to PVDF membrane (BioRad). The autoradiogram was developed for 1 week to detect [14C] acetate labeled peptides and then the PVDF membrane was stained with Coomassie blue (Sigma) to visualize peptide bands. The autoradiogram and stained membrane were overlaid to identify acetylated peptide bands. Labeled peptide bands were excised and sequenced by Edman degradation microsequencing (Protein Chemistry Laboratory, SAIC Frederick, Inc.).
Plasmids, HSP90 Subcloning, Mutagenesis, and Transfection
pcDNA3-ErbB2 was constructed as described (Tzahar et al., 1996). pcDNA3-HA-Hif1α and pcDNA3-CHIP were described previously (Isaacs et al., 2002; Xu et al., 2002). Androgen receptor expression plasmid was a kind gift from D. Smith (Mayo Clinic, Scottsdale, AZ). Aha1 cDNA, cloned into the pcDNA3 vector as a C-terminal fusion with the FLAG eptiope tag, was generously supplied by M. Yoshida (RIKEN, Saitama, kind gift from W. Houry, University of Toronto, Toronto, ON, Canada). The PCR product was subcloned into the BamHI/XhoI sites of the pcDNA3-FLAG vector or the pcDNA3-HA vector (Invitrogen). Hsp90 mutants were generated following the QuickChange mutagenesis kit protocol (Stratagene). K294 was mutated to one of 3 alternative amino acids (R, Q, or A). Sequences of both wild type and mutant Hsp90α constructs were confirmed by sequencing the entire cDNA. pcDNA3-FLAG-pcDNA3-CHIP constructs were transfected using Fugene 6 (Roche) following manufacturer’s protocols. Transfectin (Bio-Rad) was used for transfection of SKBr3 cells.
Immunoprecipitation and Immunoblotting
COS7, NIH 3T3, or SkBr3 cells were transfected as indicated. Cells were lysed (20 mM PIPES pH 7.0, 100 mM NaCl, 1 mM MgCl2, 0.1% NP40, 20 mM Na2MoO4, 2 mM Na3VO4, 30 mM NaF, and protease inhibitors) and immunoprecipitated for 3 hours at 4°C with anti-FLAG (Sigma), anti-HA (Covance), anti-p60v-src (Calbiochem), anti-p53 (Santa-Cruz), anti-FKBP52 (Stressgen), or anti-p60Hop (Stressgen). Immunopellets were washed 4 times with fresh lysis buffer and eluted with 2x SDS-PAGE sample buffer. Precipitated proteins were separated by SDS-PAGE and transferred to PVDF membranes. Co-immunoprecipitated proteins were detected by immunoblotting with indicated antibodies recognizing FLAG (Sigma), HA (Covance), ErbB2 (Neomarkers), androgen receptor, p23, Hsp70 (Stressgen), p50cdc37 (Neomarkers), or CHIP (a kind gift from Cam Patterson, University of North Carolina).
Chk1 Chaperoning Reconstitution Assay and In Vitro Co-chaperone Binding
Chk1 chaperoning and subsequent kinase assays were conducted as described by Arlander et al. (Arlander et al., 2006) using 0.7 μg of resin-bound GST-Chk1 (1-265), 1 μg of purified WT or mutant Hsp90 and the following amounts of other purified chaperone proteins: 10 μg Hsp70, 2 μg Hdj1, 2 μg p50cdc37, 0.06 Units CK2, 2.5 μg p60hop. In vitro kinase activity was calculated from the phosphorimager quantitation of the membrane-bound, radiolabeled protein substrate of the chaperoned samples compared to the unchaperoned control. Antibodies used for detecting p50cdc37 interactions were from Affinity Bioreagents; other antibodies used are as described above.
Yeast Strains, Plasmids, and Growth Conditions
The Saccharomyces cerevisiae strains used in this study are all derivatives of either ΔPLD82a or RG771 and are listed in Table 1 (Supplementary Information). The parental plasmid used to express the human isoforms of Hsp90 is based on the pESC-URA Vector (Stratagene). The URA3 marker of this high copy, 2μ-based plasmid was disrupted by HIS3 and the GAL10 promoter was replaced by a constitutive 700 bp fragment of the ADH1 promoter using in vivo homologous recombination. The resulting plasmid (pESC-URA::HIS3-ADH1p) was linearized with Sac1 and co-transformed along with the various amplified alleles of human HSP90 into strain YKR314. Plasmids were rescued from HIS+ colonies (either before or after selecting on 5-FOA depending on viability), and the cloning of the desired products of recombination was confirmed by restriction digest and sequencing of both strands. In each case a verified clone was re-transformed into Y314 and subjected to selection by 5-FOA to generate the viable strains listed in Table 1 (Supplemental Information). To generate low copy, CEN-ARS versions of each expression construct, each verified clone was digested with SnaBI and Xba and co-transformed with pRS414 (Sikorski and Hieter, 1989) digested with ScaI and PstI into strain YKR314. Plasmids were rescued from TRP+ colonies and processed exactly as described for the 2μ-HIS3 constructs. Site-directed mutagenesis (Kitagawa and Abdulle, 2002) was used to generate K274 acetylation site mutant alleles of HSP82 in plasmid pGPD1pro-HSP82-CEN-ARS-TRP1 which was rescued from strain Y82a (kindly provided by Avrom Caplan (Fang et al., 1996)). Further details of plasmid construction and yeast strains can be found in Supplemental Information. Yeast strains were grown at 30°C in YPD medium or in SD medium supplemented with appropriate amino acids and bases, and transformed with plasmids using standard procedures (Rose and Broach, 1990). SGal medium contains YNB supplemented with 2% galactose (filter sterilized) and a drop-out mixture of amino acids and bases. Five-fold serial dilutions of cells from an exponential culture were spotted onto YPD, SD, or SGAL plates and incubated at 30°C for 2-3 days or as indicated in the Figure Legends.
Supplementary Material
Acknowledgements
We thank M. Yoshida, W. Houry, D. Smith, S. Lindquist, E. Craig and A. Caplan for their generosity in supplying plasmids and yeast strains, C. Patterson for supplying anti-CHIP antibody, and T. A. J. Haystead for supplying ATP-agarose. We are also grateful to Ed Mimnaugh and Wanping Xu for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlander SJ, Felts SJ, Wagner JM, Stensgard B, Toft DO, Karnitz LM. Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J Biol Chem. 2006;281:2989–2998. doi: 10.1074/jbc.M508687200. [DOI] [PubMed] [Google Scholar]
- Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005a;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- Bali P, Pranpat M, Swaby R, Fiskus W, Yamaguchi H, Balasis M, Rocha K, Wang HG, Richon V, Bhalla K. Activity of suberoylanilide hydroxamic Acid against human breast cancer cells with amplification of her-2. Clin Cancer Res. 2005b;11:6382–6389. doi: 10.1158/1078-0432.CCR-05-0344. [DOI] [PubMed] [Google Scholar]
- Blagg BS, Kerr TD. Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med Res Rev. 2006;26:310–338. doi: 10.1002/med.20052. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Robey R, Sackett DL, Du L, Traganos F, Darzynkiewicz Z, Fojo T, Bates SE. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol Cancer Ther. 2002;1:937–941. [PubMed] [Google Scholar]
- Chadli A, Bouhouche I, Sullivan W, Stensgard B, McMahon N, Catelli MG, Toft DO. Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. Proc Natl Acad Sci U S A. 2000;97:12524–12529. doi: 10.1073/pnas.220430297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Fliss AE, Robins DM, Caplan AJ. Hsp90 regulates androgen receptor hormone binding affinity in vivo. J Biol Chem. 1996;271:28697–28702. doi: 10.1074/jbc.271.45.28697. [DOI] [PubMed] [Google Scholar]
- Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, Horinouchi S. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- Hartson SD, Thulasiraman V, Huang W, Whitesell L, Matts RL. Molybdate inhibits hsp90, induces structural changes in its C-terminal domain, and alters its interactions with substrates. Biochemistry. 1999;38:3837–3849. doi: 10.1021/bi983027s. [DOI] [PubMed] [Google Scholar]
- Huai Q, Wang H, Liu Y, Kim HY, Toft D, Ke H. Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding. Structure. 2005;13:579–590. doi: 10.1016/j.str.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. Acetylation of Estrogen Receptor Alpha by p300 at Lysines 266 and 268 Enhances the DNA Binding and Transactivation Activities of the Receptor. Mol Endocrinol. 2006 doi: 10.1210/me.2005-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Abdulle R. In vivo site-directed mutagenesis of yeast plasmids using a three-fragment homologous recombination system. Biotechniques. 2002;33 doi: 10.2144/02332bm07. passim. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Louvion JF, Abbas Terki T, Picard D. Hsp90 is required for pheromone signaling in yeast. Mol Biol Cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean MJ, Llordella MM, Bot N, Picard D. A yeast-based assay reveals a functional defect of the Q488H polymorphism in human Hsp90alpha. Biochem Biophys Res Commun. 2005;337:133–137. doi: 10.1016/j.bbrc.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Liao C, Hu B, Mark Roe S, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. Embo J. 2004;23:511–519. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimnaugh EG, Worland PJ, Whitesell L, Neckers LM. Possible role for serine/threonine phosphorylation in the regulation of the heteroprotein complex between the hsp90 stress protein and the pp60v-src tyrosine kinase. J Biol Chem. 1995;270:28654–28659. doi: 10.1074/jbc.270.48.28654. [DOI] [PubMed] [Google Scholar]
- Murphy PJ, Morishima Y, Kovacs JJ, Yao TP, Pratt WB. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. J Biol Chem. 2005;280:33792–33799. doi: 10.1074/jbc.M506997200. [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Sato N, Iwanari H, Yamashita H, Takagi T. Domain structures and immunogenic regions of the 90-kDa heat-shock protein (HSP90). Probing with a library of anti-HSP90 monoclonal antibodies and limited proteolysis. J Biol Chem. 1997;272:26179–26187. doi: 10.1074/jbc.272.42.26179. [DOI] [PubMed] [Google Scholar]
- Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, Moscinski L, Smith C, Wu J, Jove R, et al. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or -refractory chronic myelogenous leukemiablast crisis cells. Cancer Res. 2003;63:5126–5135. [PubMed] [Google Scholar]
- Ogiso H, Kagi N, Matsumoto E, Nishimoto M, Arai R, Shirouzu M, Mimura J, Fujii-Kuriyama Y, Yokoyama S. Phosphorylation analysis of 90 kDa heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry. 2004;43:15510–15519. doi: 10.1021/bi048736m. [DOI] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- Piper PW, Panaretou B, Millson SH, Trumana A, Mollapour M, Pearl LH, Prodromou C. Yeast is selectively hypersensitised to heat shock protein 90 (Hsp90)-targetting drugs with heterologous expression of the human Hsp90beta, a property that can be exploited in screens for new Hsp90 chaperone inhibitors. Gene. 2003;302:165–170. doi: 10.1016/s0378-1119(02)01102-2. [DOI] [PubMed] [Google Scholar]
- Rose AB, Broach JR. Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- Sartor O, McLellan CA, Chiueh T. Comparison of src-family cDNAs reveals distinct mechanisms underlying focus formation in transfected fibroblasts. J Biol Chem. 1992;267:21044–21051. [PubMed] [Google Scholar]
- Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A. 2000;97:10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger SK, Suhre MH, Wegele H, Buchner J. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. Embo J. 2006;25:367–376. doi: 10.1038/sj.emboj.7600930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Tsay YG, Tan BC, Lo WY, Lee SC. Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J Biol Chem. 2003;278:25568–25576. doi: 10.1074/jbc.M212574200. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci U S A. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci U S A. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- Zhao YG, Gilmore R, Leone G, Coffey MC, Weber B, Lee PW. Hsp90 phosphorylation is linked to its chaperoning function. Assembly of the reovirus cell attachment protein. J Biol Chem. 2001;276:32822–32827. doi: 10.1074/jbc.M105562200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.